A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met
- PMID: 11387228
- PMCID: PMC125478
- DOI: 10.1093/emboj/20.11.2954
A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met
Abstract
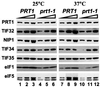
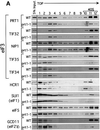
Yeast translation initiation factor 3 contains five core subunits (known as TIF32, PRT1, NIP1, TIF34 and TIF35) and a less tightly associated component known as HCR1. We found that a stable subcomplex of His8-PRT1, NIP1 and TIF32 (PN2 subcomplex) could be affinity purified from a strain overexpressing these eIF3 subunits. eIF5, eIF1 and HCR1 co-purified with this subcomplex, but not with distinct His8-PRT1- TIF34-TIF35 (P45) or His8-PRT1-TIF32 (P2) sub complexes. His8-PRT1 and NIP1 did not form a stable binary subcomplex. These results provide in vivo evidence that TIF32 bridges PRT1 and NIP1, and that eIFs 1 and 5 bind to NIP1, in native eIF3. Heat-treated prt1-1 extracts are defective for Met-tRNA(i)Met binding to 40S subunits, and we also observed defective 40S binding of mRNA, eIFs 1 and 5 and eIF3 itself in these extracts. We could rescue 40S binding of Met- tRNA(i)Met and mRNA, and translation of luciferase mRNA, in a prt1-1 extract almost as well with purified PN2 subcomplex as with five-subunit eIF3, whereas the P45 subcomplex was nearly inactive. Thus, several key functions of eIF3 can be carried out by the PRT1-TIF32-NIP1 subcomplex.
Figures
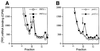





Similar articles
-
Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding.EMBO J. 2001 Feb 15;20(4):891-904. doi: 10.1093/emboj/20.4.891. EMBO J. 2001. PMID: 11179233 Free PMC article.
-
Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection.Mol Cell Biol. 2004 Nov;24(21):9437-55. doi: 10.1128/MCB.24.21.9437-9455.2004. Mol Cell Biol. 2004. PMID: 15485912 Free PMC article.
-
The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo.Genes Dev. 2003 Mar 15;17(6):786-99. doi: 10.1101/gad.1065403. Genes Dev. 2003. PMID: 12651896 Free PMC article.
-
The scanning mechanism of eukaryotic translation initiation.Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review.
-
A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition.Cold Spring Harb Symp Quant Biol. 2001;66:403-15. doi: 10.1101/sqb.2001.66.403. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762043 Review. No abstract available.
Cited by
-
CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression.Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15688-93. doi: 10.1073/pnas.0506791102. Epub 2005 Oct 14. Proc Natl Acad Sci U S A. 2005. PMID: 16227438 Free PMC article.
-
Endoplasmic reticulum stress: New insights into the pathogenesis and treatment of retinal degenerative diseases.Prog Retin Eye Res. 2020 Nov;79:100860. doi: 10.1016/j.preteyeres.2020.100860. Epub 2020 Apr 6. Prog Retin Eye Res. 2020. PMID: 32272207 Free PMC article. Review.
-
Yeast translation elongation factor eEF3 promotes late stages of tRNA translocation.EMBO J. 2021 Mar 15;40(6):e106449. doi: 10.15252/embj.2020106449. Epub 2021 Feb 8. EMBO J. 2021. PMID: 33555093 Free PMC article.
-
The eIF3 complex of Leishmania-subunit composition and mode of recruitment to different cap-binding complexes.Nucleic Acids Res. 2015 Jul 27;43(13):6222-35. doi: 10.1093/nar/gkv564. Epub 2015 Jun 19. Nucleic Acids Res. 2015. PMID: 26092695 Free PMC article.
-
Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.Nucleic Acids Res. 2015 Mar 11;43(5):2946-57. doi: 10.1093/nar/gkv053. Epub 2015 Feb 17. Nucleic Acids Res. 2015. PMID: 25690901 Free PMC article.
References
-
- Asano K., Phan,L., Anderson,J. and Hinnebusch,A.G. (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem., 273, 18573–18585. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

