Introns and their positions affect the translational activity of mRNA in plant cells
- PMID: 11375930
- PMCID: PMC1083884
- DOI: 10.1093/embo-reports/kve090
Introns and their positions affect the translational activity of mRNA in plant cells
Abstract
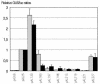
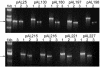
In an attempt to further increase transgene expression levels in plants over and above the enhancement obtained with a 5' untranslated leader intron, three different maize introns were inserted at three different positions within the coding sequence of the luciferase reporter gene. Constructs were transformed into maize (Black Mexican Sweet) cells and protoplasts, and their activity determined. Although all introns tested were correctly spliced, only one of them in a particular position was able to enhance gene expression. Correct splicing sites were used for intron removal and the quantity of luciferase mRNA produced did not differ significantly. These data indicate that both the position and the sequence of an intron have marked effects on expression levels, suggesting that nuclear processing of the pre-mRNA determines final expression levels through the structure of the mRNP.
Figures



Similar articles
-
Insertion of non-intron sequence into maize introns interferes with splicing.Nucleic Acids Res. 1992 Oct 11;20(19):5181-7. doi: 10.1093/nar/20.19.5181. Nucleic Acids Res. 1992. PMID: 1383942 Free PMC article.
-
The regulation of gene expression in transformed maize aleurone and endosperm protoplasts. Analysis of promoter activity, intron enhancement, and mRNA untranslated regions on expression.Plant Physiol. 1994 Nov;106(3):929-39. doi: 10.1104/pp.106.3.929. Plant Physiol. 1994. PMID: 7824660 Free PMC article.
-
Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process.Plant J. 1997 Oct;12(4):895-9. doi: 10.1046/j.1365-313x.1997.12040895.x. Plant J. 1997. PMID: 9375400
-
The enduring mystery of intron-mediated enhancement.Plant Sci. 2015 Aug;237:8-15. doi: 10.1016/j.plantsci.2015.04.017. Epub 2015 Apr 30. Plant Sci. 2015. PMID: 26089147 Review.
-
Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery.Plant Mol Biol. 1996 Oct;32(1-2):1-41. doi: 10.1007/BF00039375. Plant Mol Biol. 1996. PMID: 8980472 Review.
Cited by
-
Plants as bioreactors: Recent developments and emerging opportunities.Biotechnol Adv. 2009 Nov-Dec;27(6):811-832. doi: 10.1016/j.biotechadv.2009.06.004. Epub 2009 Jun 30. Biotechnol Adv. 2009. PMID: 19576278 Free PMC article. Review.
-
Plant Platforms for Efficient Heterologous Protein Production.Biotechnol Bioprocess Eng. 2021;26(4):546-567. doi: 10.1007/s12257-020-0374-1. Epub 2021 Aug 7. Biotechnol Bioprocess Eng. 2021. PMID: 34393545 Free PMC article. Review.
-
Relationship between polymorphism of receptor SCARB2 gene and clinical severity of enterovirus-71 associated hand-foot-mouth disease.Virol J. 2021 Jun 30;18(1):132. doi: 10.1186/s12985-021-01605-0. Virol J. 2021. PMID: 34193186 Free PMC article.
-
Using the candidate gene approach for detecting genes underlying seed oil concentration and yield in soybean.Theor Appl Genet. 2013 Jul;126(7):1839-50. doi: 10.1007/s00122-013-2096-7. Epub 2013 Apr 9. Theor Appl Genet. 2013. PMID: 23568222
-
UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants.Mol Cell Biol. 2002 Jun;22(12):4346-57. doi: 10.1128/MCB.22.12.4346-4357.2002. Mol Cell Biol. 2002. PMID: 12024044 Free PMC article.
References
-
- Callis J., Fromm, M. and Walbot, V. (1987) Introns increase gene expression in cultured maize cells. Genes Dev., 1, 1183–1200. - PubMed
-
- Evans M.J. and Scarpulla, R.C. (1989) Introns in the 3′-untranslated region can inhibit chimeric CAT and β-galactosidase gene expression. Gene, 84, 135–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

