Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa
- PMID: 11353602
- PMCID: PMC90522
- DOI: 10.1128/AAC.45.6.1615-1620.2001
Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa
Abstract
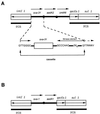
Pseudomonas aeruginosa clinical isolate SOF-1 was resistant to cefepime and susceptible to ceftazidime. This resistance phenotype was explained by the expression of OXA-31, which shared 98% amino acid identity with a class D beta-lactamase, OXA-1. The oxa-31 gene was located on a ca. 300-kb nonconjugative plasmid and on a class 1 integron. No additional efflux mechanism for cefepime was detected in P. aeruginosa SOF-1. Resistance to cefepime and susceptibility to ceftazidime in P. aeruginosa were conferred by OXA-1 as well.
Figures


Similar articles
-
In vivo selection of oxacillinase-mediated ceftazidime resistance in Pseudomonas aeruginosa.Microb Drug Resist. 2001 Fall;7(3):273-5. doi: 10.1089/10766290152652828. Microb Drug Resist. 2001. PMID: 11759089
-
Comparative in-vitro activities of newer cephalosporins cefclidin, cefepime, and cefpirome against ceftazidime- or imipenem-resistant Pseudomonas aeruginosa.J Antimicrob Chemother. 1992 Nov;30(5):633-41. doi: 10.1093/jac/30.5.633. J Antimicrob Chemother. 1992. PMID: 1493980
-
Mechanisms of resistance in clinical isolates of Pseudomonas aeruginosa less susceptible to cefepime than to ceftazidime.Clin Microbiol Infect. 2011 Dec;17(12):1817-22. doi: 10.1111/j.1469-0691.2011.03530.x. Epub 2011 May 23. Clin Microbiol Infect. 2011. PMID: 21599797
-
[New antimicrobial agent series IL: cefepime].Jpn J Antibiot. 1996 Aug;49(8):766-81. Jpn J Antibiot. 1996. PMID: 8910134 Review. Japanese. No abstract available.
-
[New antimicrobial agent series XVII: Ceftazidime].Jpn J Antibiot. 1986 Nov;39(11):2819-30. Jpn J Antibiot. 1986. PMID: 3102806 Review. Japanese. No abstract available.
Cited by
-
Resistance in Pseudomonas aeruginosa: A Narrative Review of Antibiogram Interpretation and Emerging Treatments.Antibiotics (Basel). 2023 Nov 12;12(11):1621. doi: 10.3390/antibiotics12111621. Antibiotics (Basel). 2023. PMID: 37998823 Free PMC article. Review.
-
Diversity, epidemiology, and genetics of class D beta-lactamases.Antimicrob Agents Chemother. 2010 Jan;54(1):24-38. doi: 10.1128/AAC.01512-08. Epub 2009 Aug 31. Antimicrob Agents Chemother. 2010. PMID: 19721065 Free PMC article. Review.
-
Antimicrobial Resistance in Romania: Updates on Gram-Negative ESCAPE Pathogens in the Clinical, Veterinary, and Aquatic Sectors.Int J Mol Sci. 2023 Apr 26;24(9):7892. doi: 10.3390/ijms24097892. Int J Mol Sci. 2023. PMID: 37175597 Free PMC article. Review.
-
The Prevalence of Antibiotic Resistance Phenotypes and Genotypes in Multidrug-Resistant Bacterial Isolates from the Academic Hospital of Jaén, Spain.Antibiotics (Basel). 2024 May 9;13(5):429. doi: 10.3390/antibiotics13050429. Antibiotics (Basel). 2024. PMID: 38786157 Free PMC article.
-
Antimicrobial Resistance Patterns and Prevalence of blaPER-1 and blaVEB-1 Genes Among ESBL-producing Pseudomonas aeruginosa Isolates in West of Iran.Jundishapur J Microbiol. 2014 Jan;7(1):e8888. doi: 10.5812/jjm.8888. Epub 2014 Jan 1. Jundishapur J Microbiol. 2014. PMID: 25147662 Free PMC article.
References
-
- Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

