Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development
- PMID: 11340190
- PMCID: PMC135555
- DOI: 10.1105/tpc.13.5.1179
Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development
Abstract
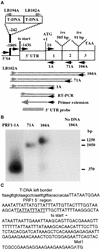
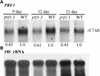
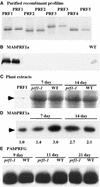
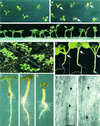
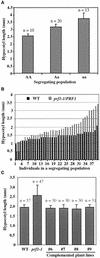
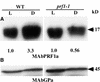
Profilin (PRF) is a low-molecular-weight actin binding protein encoded by a diverse gene family in plants. Arabidopsis PRF1 transcripts are moderately well expressed in all vegetative organs. A regulatory mutant in PRF1, prf1-1, was isolated from a library of T-DNA insertions. The insertion disrupted the promoter region of PRF1 100 bp upstream from the transcriptional start site. Although steady state levels of PRF1 transcripts appeared normal in mature prf1-1 plants, the levels in young seedlings were only one-half those observed in wild type. Reactions with a PRF1 isovariant-specific monoclonal antiserum and general anti-profilin antisera demonstrated that PRF1 protein levels also were one-half those found in wild-type seedlings, although total profilin levels were unaffected. Mutant seedlings no longer could downregulate PRF1 levels in the light, as did wild type. Consistent with their molecular phenotypes, young mutant seedlings displayed several morphological phenotypes but developed into apparently normal adult plants. Their initial germination rate and development were slow, and they produced excessive numbers of root hairs. Mutant seedlings had abnormally raised cotyledons, elongated hypocotyls, and elongated cells in the hypocotyl, typical of phenotypes associated with some defects in light and circadian responses. A wild-type PRF1 transgene fully complements the hypocotyl phenotypes in the prf1-1 mutant. The ability of profilin to regulate actin polymerization and participate directly in signal transduction pathways is discussed in light of the prf1-1 phenotypes.
Figures
Similar articles
-
Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis.Plant Physiol. 2000 Dec;124(4):1637-47. doi: 10.1104/pp.124.4.1637. Plant Physiol. 2000. PMID: 11115881 Free PMC article.
-
Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin.Plant J. 1996 Aug;10(2):269-79. doi: 10.1046/j.1365-313x.1996.10020269.x. Plant J. 1996. PMID: 8771785
-
The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes.Plant Physiol. 1996 May;111(1):115-26. doi: 10.1104/pp.111.1.115. Plant Physiol. 1996. PMID: 8685262 Free PMC article.
-
Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues.Cell Motil Cytoskeleton. 2002 May;52(1):22-32. doi: 10.1002/cm.10029. Cell Motil Cytoskeleton. 2002. PMID: 11977080
-
[Profilins in plant cells].Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2006 Jun;32(3):261-70. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2006. PMID: 16775392 Review. Chinese.
Cited by
-
The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis.Plant Physiol. 2003 Aug;132(4):2034-44. doi: 10.1104/pp.103.028563. Plant Physiol. 2003. PMID: 12913159 Free PMC article.
-
GLABRA2 Regulates Actin Bundling Protein VILLIN1 in Root Hair Growth in Response to Osmotic Stress.Plant Physiol. 2020 Sep;184(1):176-193. doi: 10.1104/pp.20.00480. Epub 2020 Jul 7. Plant Physiol. 2020. PMID: 32636342 Free PMC article.
-
Photomorphogenesis.Arabidopsis Book. 2002;1:e0054. doi: 10.1199/tab.0054. Epub 2002 Aug 12. Arabidopsis Book. 2002. PMID: 22303211 Free PMC article. No abstract available.
-
The Arabidopsis cytoskeletal genome.Arabidopsis Book. 2003;2:e0096. doi: 10.1199/tab.0096. Epub 2003 Sep 30. Arabidopsis Book. 2003. PMID: 22303225 Free PMC article.
-
Overexpression of profilin 3 affects cell elongation and F-actin organization in Arabidopsis thaliana.Plant Cell Rep. 2013 Jan;32(1):149-60. doi: 10.1007/s00299-012-1349-2. Epub 2012 Oct 4. Plant Cell Rep. 2013. PMID: 23052593
References
-
- Calman, B.G., and Chamberlain, S.C. (1992). Localization of actin filaments and microtubules in the cells of the Limulus lateral and ventral eyes. Vis. Neurosci. 9 365–375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

