Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis
- PMID: 11333912
- PMCID: PMC114936
- DOI: 10.1128/JVI.75.11.5315-5327.2001
Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis
Abstract
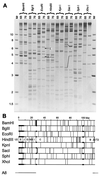
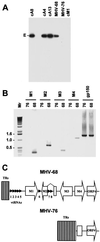
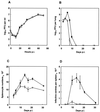
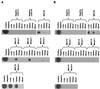
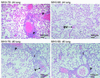
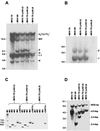
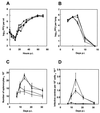
Infection of mice by murine gammaherpesvirus 68 (MHV-68) is an excellent small-animal model of gammaherpesvirus pathogenesis in a natural host. We have carried out comparative studies of another herpesvirus, murine herpesvirus 76 (MHV-76), which was isolated at the same time as MHV-68 but from a different murid host, the yellow-necked mouse (Apodemus flavicollis). Molecular analyses revealed that the MHV-76 genome is essentially identical to that of MHV-68, except for deletion of 9,538 bp at the left end of the unique region. MHV-76 is therefore a deletion mutant that lacks four genes unique to MHV-68 (M1, M2, M3, and M4) as well as the eight viral tRNA-like genes. Replication of MHV-76 in cell culture was identical to that of MHV-68. However, following infection of mice, MHV-76 was cleared more rapidly from the lungs. In line with this, there was an increased inflammatory response in lungs with MHV-76. Splenomegaly was also significantly reduced following MHV-76 infection, and much less latent MHV-76 was detected in the spleen. Nevertheless, MHV-76 maintained long-term latency in the lungs and spleen. We utilized a cosmid containing the left end of the MHV-68 genome to reinsert the deleted sequence into MHV-76 by recombination in infected cells, and we isolated a rescuant virus designated MHV-76(cA8+)4 which was ostensibly genetically identical to MHV-68. The growth properties of the rescuant in infected mice were identical to those of MHV-68. These results demonstrate that genetic elements at the left end of the unique region of the MHV-68 genome play vital roles in host evasion and are critical to the development of splenic pathology.
Figures
Similar articles
-
Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency.J Virol. 2002 Jul;76(13):6532-44. doi: 10.1128/jvi.76.13.6532-6544.2002. J Virol. 2002. PMID: 12050366 Free PMC article.
-
The m4 gene of murine gammaherpesvirus modulates productive and latent infection in vivo.J Virol. 2004 Jan;78(2):758-67. doi: 10.1128/jvi.78.2.758-767.2004. J Virol. 2004. PMID: 14694108 Free PMC article.
-
The M4 gene of murine gammaherpesvirus 68 modulates latent infection.J Gen Virol. 2006 Apr;87(Pt 4):803-807. doi: 10.1099/vir.0.81577-0. J Gen Virol. 2006. PMID: 16528028
-
Murine herpesvirus pathogenesis: a model for the analysis of molecular mechanisms of human gamma herpesvirus infections.Acta Microbiol Immunol Hung. 2005;52(1):41-71. doi: 10.1556/AMicr.52.2005.1.2. Acta Microbiol Immunol Hung. 2005. PMID: 15957234 Review.
-
Natural history of murine gamma-herpesvirus infection.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):569-79. doi: 10.1098/rstb.2000.0779. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313012 Free PMC article. Review.
Cited by
-
Murine gammaherpesvirus (MHV) MK3 gene sequence diversity among 72, 4556, and 68 strains.Virus Genes. 2006 Aug;33(1):51-8. doi: 10.1007/s11262-005-0038-4. Virus Genes. 2006. PMID: 16791419
-
Role of tachykinins in the host response to murine gammaherpesvirus infection.J Virol. 2001 Nov;75(21):10467-71. doi: 10.1128/JVI.75.21.10467-10471.2001. J Virol. 2001. PMID: 11581415 Free PMC article.
-
Herpesvirus microRNAs: phenotypes and functions.Curr Opin Virol. 2011 Sep;1(3):211-5. doi: 10.1016/j.coviro.2011.04.003. Curr Opin Virol. 2011. PMID: 21927637 Free PMC article. Review.
-
Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo.mBio. 2015 Feb 17;6(1):e01670-14. doi: 10.1128/mBio.01670-14. mBio. 2015. PMID: 25691585 Free PMC article.
-
Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency.J Virol. 2002 Jul;76(13):6532-44. doi: 10.1128/jvi.76.13.6532-6544.2002. J Virol. 2002. PMID: 12050366 Free PMC article.
References
-
- Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. - PubMed
-
- Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

