Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2
- PMID: 11312322
- PMCID: PMC114205
- DOI: 10.1128/JVI.75.10.4519-4527.2001
Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2
Abstract
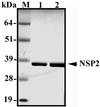
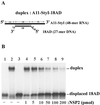
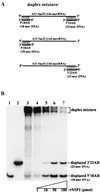
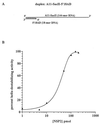
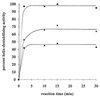
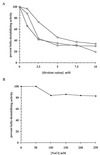
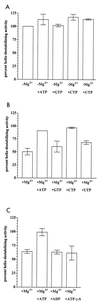
The rotavirus nonstructural protein NSP2 self-assembles into homomultimers, binds single-stranded RNA nonspecifically, possesses a Mg2+-dependent nucleoside triphosphatase (NTPase) activity, and is a component of replication intermediates. Because these properties are characteristics of known viral helicases, we examined the possibility that this was also an activity of NSP2 by using a strand displacement assay and purified bacterially expressed protein. The results revealed that, under saturating concentrations, NSP2 disrupted both DNA-RNA and RNA-RNA duplexes; hence, the protein possesses helix-destabilizing activity. However, unlike typical helicases, NSP2 required neither a divalent cation nor a nucleotide energy source for helix destabilization. Further characterization showed that NSP2 displayed no polarity in destabilizing a partial duplex. In addition, helix destabilization by NSP2 was found to proceed cooperatively and rapidly. The presence of Mg2+ and other divalent cations inhibited by approximately one-half the activity of NSP2, probably due to the increased stability of the duplex substrate brought on by the cations. In contrast, under conditions where NSP2 functions as an NTPase, its helix-destabilizing activity was less sensitive to the presence of Mg2+, suggesting that in the cellular environment the two activities associated with the protein, helix destabilization and NTPase, may function together. Although distinct from typical helicases, the helix-destabilizing activity of NSP2 is quite similar to that of the sigmaNS protein of reovirus and to the single-stranded DNA-binding proteins (SSBs) involved in double-stranded DNA replication. The presence of SSB-like nonstructural proteins in two members of the family Reoviridae suggests a common mechanism of unwinding viral mRNA prior to packaging and subsequent minus-strand RNA synthesis.
Figures


Similar articles
-
Rotavirus NSP2 interferes with the core lattice protein VP2 in initiation of minus-strand synthesis.Virology. 2003 Aug 15;313(1):261-73. doi: 10.1016/s0042-6822(03)00302-7. Virology. 2003. PMID: 12951038
-
Multimers of the bluetongue virus nonstructural protein, NS2, possess nucleotidyl phosphatase activity: similarities between NS2 and rotavirus NSP2.Virology. 2001 Feb 15;280(2):221-31. doi: 10.1006/viro.2000.0764. Virology. 2001. PMID: 11162836
-
Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae.Virus Res. 2004 Apr;101(1):57-66. doi: 10.1016/j.virusres.2003.12.006. Virus Res. 2004. PMID: 15010217 Review.
-
Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes.J Biol Chem. 2001 Mar 30;276(13):9679-87. doi: 10.1074/jbc.M009398200. Epub 2000 Dec 19. J Biol Chem. 2001. PMID: 11121414
-
Nucleotide triphosphatase/helicase of hepatitis C virus as a target for antiviral therapy.Antiviral Res. 2002 Sep;55(3):397-412. doi: 10.1016/s0166-3542(02)00096-7. Antiviral Res. 2002. PMID: 12206878 Review.
Cited by
-
Cryo-EM structure of rotavirus B NSP2 reveals its unique tertiary architecture.J Virol. 2024 Mar 19;98(3):e0166023. doi: 10.1128/jvi.01660-23. Epub 2024 Feb 29. J Virol. 2024. PMID: 38421167 Free PMC article.
-
Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly.Viruses. 2024 May 21;16(6):814. doi: 10.3390/v16060814. Viruses. 2024. PMID: 38932107 Free PMC article. Review.
-
Viroplasms: Assembly and Functions of Rotavirus Replication Factories.Viruses. 2021 Jul 12;13(7):1349. doi: 10.3390/v13071349. Viruses. 2021. PMID: 34372555 Free PMC article. Review.
-
Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection.J Virol. 2003 May;77(10):5948-63. doi: 10.1128/jvi.77.10.5948-5963.2003. J Virol. 2003. PMID: 12719587 Free PMC article.
-
Understanding the penetrance of intrinsic protein disorder in rotavirus proteome.Int J Biol Macromol. 2020 Feb 1;144:892-908. doi: 10.1016/j.ijbiomac.2019.09.166. Epub 2019 Nov 15. Int J Biol Macromol. 2020. PMID: 31739058 Free PMC article.
References
-
- Brookes S M, Hyatt A D, Eaton B T. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J Gen Virol. 1993;74:525–530. - PubMed
-
- Chen D, Gombold J L, Ramig R F. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology. 1990;178:143–151. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

