Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole
- PMID: 11309418
- PMCID: PMC2169458
- DOI: 10.1083/jcb.153.2.381
Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole
Abstract
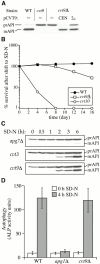
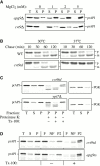
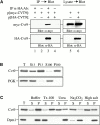
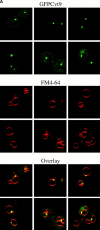
Three overlapping pathways mediate the transport of cytoplasmic material to the vacuole in Saccharomyces cerevisiae. The cytoplasm to vacuole targeting (Cvt) pathway transports the vacuolar hydrolase, aminopeptidase I (API), whereas pexophagy mediates the delivery of excess peroxisomes for degradation. Both the Cvt and pexophagy pathways are selective processes that specifically recognize their cargo. In contrast, macroautophagy nonselectively transports bulk cytosol to the vacuole for recycling. Most of the import machinery characterized thus far is required for all three modes of transport. However, unique features of each pathway dictate the requirement for additional components that differentiate these pathways from one another, including at the step of specific cargo selection.We have identified Cvt9 and its Pichia pastoris counterpart Gsa9. In S. cerevisiae, Cvt9 is required for the selective delivery of precursor API (prAPI) to the vacuole by the Cvt pathway and the targeted degradation of peroxisomes by pexophagy. In P. pastoris, Gsa9 is required for glucose-induced pexophagy. Significantly, neither Cvt9 nor Gsa9 is required for starvation-induced nonselective transport of bulk cytoplasmic cargo by macroautophagy. The deletion of CVT9 destabilizes the binding of prAPI to the membrane and analysis of a cvt9 temperature-sensitive mutant supports a direct role of Cvt9 in transport vesicle formation. Cvt9 oligomers peripherally associate with a novel, perivacuolar membrane compartment and interact with Apg1, a Ser/Thr kinase essential for both the Cvt pathway and autophagy. In P. pastoris Gsa9 is recruited to concentrated regions on the vacuole membrane that contact peroxisomes in the process of being engulfed by pexophagy. These biochemical and morphological results demonstrate that Cvt9 and the P. pastoris homologue Gsa9 may function at the step of selective cargo sequestration.
Figures



Similar articles
-
Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris.Mol Biol Cell. 2001 Dec;12(12):3821-38. doi: 10.1091/mbc.12.12.3821. Mol Biol Cell. 2001. PMID: 11739783 Free PMC article.
-
Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway.Mol Cell. 2001 Jun;7(6):1131-41. doi: 10.1016/s1097-2765(01)00263-5. Mol Cell. 2001. PMID: 11430817 Free PMC article.
-
Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway.J Biol Chem. 2004 Jul 16;279(29):29889-94. doi: 10.1074/jbc.M404399200. Epub 2004 May 11. J Biol Chem. 2004. PMID: 15138258 Free PMC article.
-
Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells.Annu Rev Biochem. 2000;69:303-42. doi: 10.1146/annurev.biochem.69.1.303. Annu Rev Biochem. 2000. PMID: 10966461 Review.
-
Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
Cited by
-
Atg32-dependent mitophagy sustains spermidine and nitric oxide required for heat-stress tolerance in Saccharomycescerevisiae.J Cell Sci. 2021 Jun 1;134(11):jcs253781. doi: 10.1242/jcs.253781. Epub 2021 Jun 7. J Cell Sci. 2021. PMID: 34096604 Free PMC article.
-
The Cvt pathway as a model for selective autophagy.FEBS Lett. 2010 Apr 2;584(7):1359-66. doi: 10.1016/j.febslet.2010.02.013. Epub 2010 Feb 8. FEBS Lett. 2010. PMID: 20146925 Free PMC article. Review.
-
Autophagy and the degradation of mitochondria.Mitochondrion. 2010 Jun;10(4):309-15. doi: 10.1016/j.mito.2010.01.005. Epub 2010 Jan 18. Mitochondrion. 2010. PMID: 20083234 Free PMC article. Review.
-
The molecular machinery of autophagy: unanswered questions.J Cell Sci. 2005 Jan 1;118(Pt 1):7-18. doi: 10.1242/jcs.01620. J Cell Sci. 2005. PMID: 15615779 Free PMC article.
-
Peroxisome size provides insights into the function of autophagy-related proteins.Mol Biol Cell. 2009 Sep;20(17):3828-39. doi: 10.1091/mbc.e09-03-0221. Epub 2009 Jul 15. Mol Biol Cell. 2009. PMID: 19605559 Free PMC article.
References
-
- Baba M., Osumi M., Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

