Treble clef finger--a functionally diverse zinc-binding structural motif
- PMID: 11292843
- PMCID: PMC31318
- DOI: 10.1093/nar/29.8.1703
Treble clef finger--a functionally diverse zinc-binding structural motif
Abstract
Detection of similarity is particularly difficult for small proteins and thus connections between many of them remain unnoticed. Structure and sequence analysis of several metal-binding proteins reveals unexpected similarities in structural domains classified as different protein folds in SCOP and suggests unification of seven folds that belong to two protein classes. The common motif, termed treble clef finger in this study, forms the protein structural core and is 25-45 residues long. The treble clef motif is assembled around the central zinc ion and consists of a zinc knuckle, loop, beta-hairpin and an alpha-helix. The knuckle and the first turn of the helix each incorporate two zinc ligands. Treble clef domains constitute the core of many structures such as ribosomal proteins L24E and S14, RING fingers, protein kinase cysteine-rich domains, nuclear receptor-like fingers, LIM domains, phosphatidylinositol-3-phosphate-binding domains and His-Me finger endonucleases. The treble clef finger is a uniquely versatile motif adaptable for various functions. This small domain with a 25 residue structural core can accommodate eight different metal-binding sites and can have many types of functions from binding of nucleic acids, proteins and small molecules, to catalysis of phosphodiester bond hydrolysis. Treble clef motifs are frequently incorporated in larger structures or occur in doublets. Present analysis suggests that the treble clef motif defines a distinct structural fold found in proteins with diverse functional properties and forms one of the major zinc finger groups.
Figures
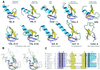

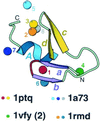
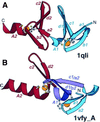
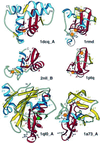
Similar articles
-
Cooperative metal binding and helical folding in model peptides of treble-clef zinc fingers.Chemistry. 2009;15(19):4798-810. doi: 10.1002/chem.200900147. Chemistry. 2009. PMID: 19388025
-
Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system.Mol Biosyst. 2011 Jul;7(7):2261-77. doi: 10.1039/c1mb05061c. Epub 2011 May 6. Mol Biosyst. 2011. PMID: 21547297 Free PMC article.
-
The UBR-box and its relationship to binuclear RING-like treble clef zinc fingers.Biol Direct. 2015 Jul 17;10:36. doi: 10.1186/s13062-015-0066-5. Biol Direct. 2015. PMID: 26185100 Free PMC article.
-
Zinc fingers--folds for many occasions.IUBMB Life. 2002 Dec;54(6):351-5. doi: 10.1080/15216540216035. IUBMB Life. 2002. PMID: 12665246 Review.
-
New types of conserved sequence domains in DNA-binding regions of homing endonucleases.Trends Biochem Sci. 2003 Sep;28(9):473-7. doi: 10.1016/S0968-0004(03)00170-1. Trends Biochem Sci. 2003. PMID: 13678957 Review.
Cited by
-
Zinc finger structure determination by NMR: Why zinc fingers can be a handful.Prog Nucl Magn Reson Spectrosc. 2022 Jun-Aug;130-131:62-105. doi: 10.1016/j.pnmrs.2022.07.001. Epub 2022 Jul 15. Prog Nucl Magn Reson Spectrosc. 2022. PMID: 36113918 Free PMC article. Review.
-
Probing the determinants of diacylglycerol binding affinity in the C1B domain of protein kinase Cα.J Mol Biol. 2011 May 20;408(5):949-70. doi: 10.1016/j.jmb.2011.03.020. Epub 2011 Mar 17. J Mol Biol. 2011. PMID: 21419781 Free PMC article.
-
Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex.EMBO J. 2004 Sep 15;23(18):3589-98. doi: 10.1038/sj.emboj.7600376. Epub 2004 Sep 2. EMBO J. 2004. PMID: 15343268 Free PMC article.
-
The structure of Jann_2411 (DUF1470) from Jannaschia sp. at 1.45 Å resolution reveals a new fold (the ABATE domain) and suggests its possible role as a transcription regulator.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Oct 1;66(Pt 10):1198-204. doi: 10.1107/S1744309109025196. Epub 2009 Oct 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20944211 Free PMC article.
-
Trans-editing of mischarged tRNAs.Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15422-7. doi: 10.1073/pnas.2136934100. Epub 2003 Dec 8. Proc Natl Acad Sci U S A. 2003. PMID: 14663147 Free PMC article.
References
-
- Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. - PubMed
-
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. - PubMed
-
- Teplyakov A., Polyakov,K., Obmolova,G., Strokopytov,B., Kuranova,I., Osterman,A., Grishin,N., Smulevitch,S., Zagnitko,O., Galperina,O. et al. (1992) Crystal structure of carboxypeptidase T from Thermoactinomyces vulgaris. Eur. J. Biochem., 208, 281–288. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

