Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae
- PMID: 11292809
- PMCID: PMC99506
- DOI: 10.1128/JB.183.9.2881-2887.2001
Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae
Abstract
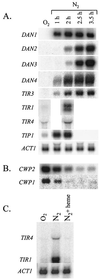
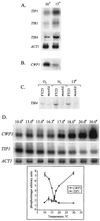
The DAN/TIR genes encode nine cell wall mannoproteins in Saccharomyces cerevisiae which are expressed during anaerobiosis (DAN1, DAN2, DAN3, DAN4, TIR1, TIR2, TIR3, TIR4, and TIP1). Most are expressed within an hour of an anaerobic shift, but DAN2 and DAN3 are expressed after about 3 h. At the same time, CWP1 and CWP2, the genes encoding the major mannoproteins, are down-regulated, suggesting that there is a programmed remodeling of the cell wall in which Cwp1 and Cwp2 are replaced by nine anaerobic counterparts. TIP1, TIR1, TIR2, and TIR4 are also induced during cold shock. Correspondingly, CWP1 is down-regulated during cold shock. As reported elsewhere, Mox4 is a heme-inhibited activator, and Mot3 is a heme-induced repressor of the DAN/TIR genes (but not of TIP1). We show that CWP2 (but not CWP1) is controlled by the same factors, but in reverse fashion-primarily by Mot3 (which can function as either an activator or repressor) but also by Mox4, accounting for the reciprocal regulation of the two groups of genes. Disruptions of TIR1, TIR3, or TIR4 prevent anaerobic growth, indicating that each protein is essential for anaerobic adaptation. The Dan/Tir and Cwp proteins are homologous, with the greatest similarities shown within three subgroups: the Dan proteins, the Tip and Tir proteins, and, more distantly, the Cwp proteins. The clustering of homology corresponds to differences in expression: the Tip and Tir proteins are expressed during hypoxia and cold shock, the Dan proteins are more stringently repressed by oxygen and insensitive to cold shock, and the Cwp proteins are oppositely regulated by oxygen and temperature.
Figures



Similar articles
-
Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites.Nucleic Acids Res. 2001 Feb 1;29(3):799-808. doi: 10.1093/nar/29.3.799. Nucleic Acids Res. 2001. PMID: 11160904 Free PMC article.
-
Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae.Genetics. 2001 Mar;157(3):1169-77. doi: 10.1093/genetics/157.3.1169. Genetics. 2001. PMID: 11238402 Free PMC article.
-
Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast.Mol Cell Biol. 2007 Mar;27(6):2037-47. doi: 10.1128/MCB.02297-06. Epub 2007 Jan 8. Mol Cell Biol. 2007. PMID: 17210643 Free PMC article.
-
Rox1 mediated repression. Oxygen dependent repression in yeast.Adv Exp Med Biol. 2000;475:185-95. Adv Exp Med Biol. 2000. PMID: 10849660 Review.
-
Cell Surface Interference with Plasma Membrane and Transport Processes in Yeasts.Adv Exp Med Biol. 2016;892:11-31. doi: 10.1007/978-3-319-25304-6_2. Adv Exp Med Biol. 2016. PMID: 26721269 Review.
Cited by
-
Activator and repressor functions of the Mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae.Eukaryot Cell. 2013 May;12(5):636-47. doi: 10.1128/EC.00037-13. Epub 2013 Feb 22. Eukaryot Cell. 2013. PMID: 23435728 Free PMC article.
-
Many Saccharomyces cerevisiae Cell Wall Protein Encoding Genes Are Coregulated by Mss11, but Cellular Adhesion Phenotypes Appear Only Flo Protein Dependent.G3 (Bethesda). 2012 Jan;2(1):131-41. doi: 10.1534/g3.111.001644. Epub 2012 Jan 1. G3 (Bethesda). 2012. PMID: 22384390 Free PMC article.
-
Functional Divergence in a Multi-gene Family Is a Key Evolutionary Innovation for Anaerobic Growth in Saccharomyces cerevisiae.Mol Biol Evol. 2022 Oct 7;39(10):msac202. doi: 10.1093/molbev/msac202. Mol Biol Evol. 2022. PMID: 36134526 Free PMC article.
-
A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae.Genetics. 2006 Sep;174(1):191-201. doi: 10.1534/genetics.106.059964. Epub 2006 Jun 18. Genetics. 2006. PMID: 16783004 Free PMC article.
-
Global screening of genes essential for growth in high-pressure and cold environments: searching for basic adaptive strategies using a yeast deletion library.Genetics. 2008 Feb;178(2):851-72. doi: 10.1534/genetics.107.083063. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245339 Free PMC article.
References
-
- Cabib E, Drgon T, Drgonova J, Ford R A, Kollar R. The yeast cell wall, a dynamic structure engaged in growth and morphogenesis. Biochem Soc Trans. 1997;25:200–204. - PubMed
-
- Caro L H P, Smits G J, Van Egmond P, Chapman J W, Klis F M. Transcription of multiple cell wall protein-encoding genes in Saccharomyces cerevisiae is differentially regulated during the cell cycle. FEMS Microbiol Lett. 1998;161:345–349. - PubMed
-
- Caro L H P, Tettelin H, Vossen J H, Ram A F J, Van den Ende H, Klis F M. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

