Sequence requirements for interaction of human herpesvirus 7 origin binding protein with the origin of lytic replication
- PMID: 11264381
- PMCID: PMC114883
- DOI: 10.1128/JVI.75.8.3925-3936.2001
Sequence requirements for interaction of human herpesvirus 7 origin binding protein with the origin of lytic replication
Abstract
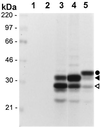
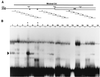
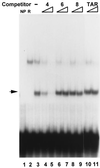
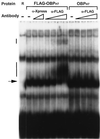
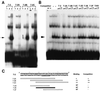
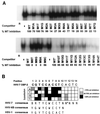
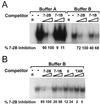
As do human herpesvirus 6 variants A and B (HHV-6A and -6B), HHV-7 encodes a homolog of the alphaherpesvirus origin binding protein (OBP), which binds at sites in the origin of lytic replication (oriLyt) to initiate DNA replication. In this study, we sought to characterize the interaction of the HHV-7 OBP (OBP(H7)) with its cognate sites in the 600-bp HHV-7 oriLyt. We expressed the carboxyl-terminal domain of OBP(H7) and found that amino acids 484 to 787 of OBP(H7) were sufficient for DNA binding activity by electrophoretic mobility shift analysis. OBP(H7) has one high-affinity binding site (OBP-2) located on one flank of an AT-rich spacer element and a low-affinity site (OBP-1) on the other. This is in contrast to the HHV-6B OBP (OBP(H6B)), which binds with similar affinity to its two cognate OBP sites in the HHV-6B oriLyt. The minimal recognition element of the OBP-2 site was mapped to a 14-bp sequence. The OBP(H7) consensus recognition sequence of the 9-bp core, BRTYCWCCT (where B is a T, G, or C; R is a G or A; Y is a T or C; and W is a T or A), overlaps with the OBP(H6B) consensus YGWYCWCCY and establishes YCWCC as the roseolovirus OBP core recognition sequence. Heteroduplex analysis suggests that OBP(H7) interacts along one face of the DNA helix, with the major groove, as do OBP(H6B) and herpes simplex virus type 1 OBP. Together, these results illustrate both conserved and divergent DNA binding properties between OBP(H7) and OBP(H6B).
Figures


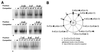
Similar articles
-
Differences in DNA binding specificity among Roseolovirus origin binding proteins.Virology. 2001 Sep 15;288(1):145-53. doi: 10.1006/viro.2001.1066. Virology. 2001. PMID: 11543667
-
The DNA binding domain of herpes simplex virus type 1 origin binding protein is a transdominant inhibitor of virus replication.Virology. 1993 Mar;193(1):73-9. doi: 10.1006/viro.1993.1104. Virology. 1993. PMID: 8382413
-
Molecular biology of human herpesviruses 6A and 6B.Infect Agents Dis. 1993 Dec;2(6):343-60. Infect Agents Dis. 1993. PMID: 8012736 Review.
-
Human herpesvirus 6B origin-binding protein: DNA-binding domain and consensus binding sequence.J Virol. 1995 Aug;69(8):4619-27. doi: 10.1128/JVI.69.8.4619-4627.1995. J Virol. 1995. PMID: 7609026 Free PMC article.
-
[Molecular biology of human herpesvirus 6: DNA replication and trans-activator genes].Nihon Rinsho. 1998 Jan;56(1):50-5. Nihon Rinsho. 1998. PMID: 9465664 Review. Japanese.
Cited by
-
Complete Unique Genome Sequence, Expression Profile, and Salivary Gland Tissue Tropism of the Herpesvirus 7 Homolog in Pigtailed Macaques.J Virol. 2016 Jul 11;90(15):6657-6674. doi: 10.1128/JVI.00651-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170755 Free PMC article.
-
Roseolovirus molecular biology: recent advances.Curr Opin Virol. 2014 Dec;9:170-7. doi: 10.1016/j.coviro.2014.10.004. Epub 2014 Nov 27. Curr Opin Virol. 2014. PMID: 25437229 Free PMC article. Review.
-
Stepwise evolution of the herpes simplex virus origin binding protein and origin of replication.J Biol Chem. 2009 Jun 12;284(24):16246-16255. doi: 10.1074/jbc.M807551200. Epub 2009 Apr 7. J Biol Chem. 2009. PMID: 19351883 Free PMC article.
-
Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt.J Virol. 2004 Nov;78(21):11664-77. doi: 10.1128/JVI.78.21.11664-11677.2004. J Virol. 2004. PMID: 15479808 Free PMC article.
References
-
- Aslani A, Simonsson S, Elias P. A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin binding protein. J Biol Chem. 2000;275:5880–5887. - PubMed
-
- Black J B, Burns D A, Goldsmith C S, Feorino P M, Kite-Powell K, Schinazi R F, Krug P W, Pellett P E. Biologic properties of human herpesvirus 7 strain SB. Virus Res. 1997;52:25–41. - PubMed
-
- Black J B, Pellett P E. Human herpesvirus 7. Rev Med Virol. 1999;9:245–262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

