Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9
- PMID: 11264375
- PMCID: PMC114877
- DOI: 10.1128/JVI.75.8.3859-3872.2001
Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9
Abstract
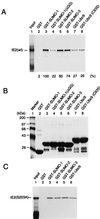
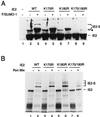
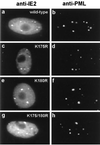
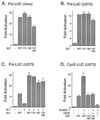
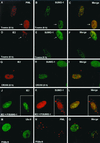
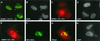
The human cytomegalovirus (HCMV) major immediate-early protein IE2 is a nuclear phosphoprotein that is believed to be a key regulator in both lytic and latent infections. Using yeast two-hybrid screening, small ubiquitin-like modifiers (SUMO-1, SUMO-2, and SUMO-3) and a SUMO-conjugating enzyme (Ubc9) were isolated as IE2-interacting proteins. In vitro binding assays with glutathione S-transferase (GST) fusion proteins provided evidence for direct protein-protein interaction. Mapping data showed that the C-terminal end of SUMO-1 is critical for interaction with IE2 in both yeast and in vitro binding assays. IE2 was efficiently modified by SUMO-1 or SUMO-2 in cotransfected cells and in cells infected with a recombinant adenovirus expressing HCMV IE2, although the level of modification was much lower in HCMV-infected cells. Two lysine residues at positions 175 and 180 were mapped as major alternative SUMO-1 conjugation sites in both cotransfected cells and an in vitro sumoylation assay and could be conjugated by SUMO-1 simultaneously. Although mutations of these lysine residues did not interfere with the POD (or ND10) targeting of IE2, overexpression of SUMO-1 enhanced IE2-mediated transactivation in a promoter-dependent manner in reporter assays. Interestingly, many other cellular proteins identified as IE2 interaction partners in yeast two-hybrid assays also interact with SUMO-1, suggesting that either directly bound or covalently conjugated SUMO moieties may act as a bridge for interactions between IE2 and other SUMO-1-modified or SUMO-1-interacting proteins. When we investigated the intracellular localization of SUMO-1 in HCMV-infected cells, the pattern changed from nuclear punctate to predominantly nuclear diffuse in an IE1-dependent manner at very early times after infection, but with some SUMO-1 protein now associated with IE2 punctate domains. However, at late times after infection, SUMO-1 was predominantly detected within viral DNA replication compartments containing IE2. Taken together, these results show that HCMV infection causes the redistribution of SUMO-1 and that IE2 both physically binds to and is covalently modified by SUMO moieties, suggesting possible modulation of both the function of SUMO-1 and protein-protein interactions of IE2 during HCMV infection.
Figures



Similar articles
-
Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b.J Virol. 2000 Mar;74(6):2510-24. doi: 10.1128/jvi.74.6.2510-2524.2000. J Virol. 2000. PMID: 10684265 Free PMC article.
-
The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10).J Virol. 1999 Dec;73(12):10458-71. doi: 10.1128/JVI.73.12.10458-10471.1999. J Virol. 1999. PMID: 10559364 Free PMC article.
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells.J Virol. 1997 Jun;71(6):4599-613. doi: 10.1128/JVI.71.6.4599-4613.1997. J Virol. 1997. PMID: 9151854 Free PMC article.
-
Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression.J Virol. 2001 Nov;75(22):10683-95. doi: 10.1128/JVI.75.22.10683-10695.2001. J Virol. 2001. PMID: 11602710 Free PMC article.
Cited by
-
SUMO modifies GβL and mediates mTOR signaling.J Biol Chem. 2024 Apr;300(4):105778. doi: 10.1016/j.jbc.2024.105778. Epub 2024 Feb 21. J Biol Chem. 2024. PMID: 38395307 Free PMC article.
-
Virus-host protein interactions as footprints of human cytomegalovirus replication.Curr Opin Virol. 2022 Feb;52:135-147. doi: 10.1016/j.coviro.2021.11.016. Epub 2021 Dec 16. Curr Opin Virol. 2022. PMID: 34923282 Free PMC article. Review.
-
Site-specific SUMOylation of viral polymerase processivity factor: a way of localizingtoND10 subnuclear domains for restricted and self-controlled reproduction of herpesvirus.Virulence. 2021 Dec;12(1):2883-2901. doi: 10.1080/21505594.2021.2000689. Virulence. 2021. PMID: 34747321 Free PMC article.
-
Sumoylation of the Carboxy-Terminal of Human Cytomegalovirus DNA Polymerase Processivity Factor UL44 Attenuates Viral DNA Replication.Front Microbiol. 2021 Apr 21;12:652719. doi: 10.3389/fmicb.2021.652719. eCollection 2021. Front Microbiol. 2021. PMID: 33967989 Free PMC article.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
References
-
- Ahn J H, Chiou C J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. - PubMed
-
- Ahn J H, Hayward G S. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

