Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs
- PMID: 11238889
- PMCID: PMC86698
- DOI: 10.1128/MCB.21.5.1515-1530.2001
Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs
Abstract
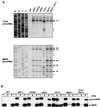
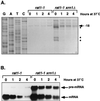
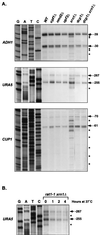
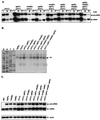
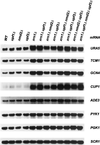
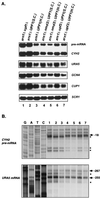
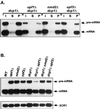
In Saccharomyces cerevisiae, rapid degradation of nonsense-containing mRNAs requires the decapping enzyme Dcp1p, the 5'-to-3' exoribonuclease Xrn1p, and the three nonsense-mediated mRNA decay (NMD) factors, Upf1p, Nmd2p, and Upf3p. To identify specific functions for the NMD factors, we analyzed the mRNA decay phenotypes of yeast strains containing deletions of DCP1 or XRN1 and UPF1, NMD2, or UPF3. Our results indicate that Upf1p, Nmd2p, and Upf3p regulate decapping and exonucleolytic degradation of nonsense-containing mRNAs. In addition, we show that these factors also regulate the same processes in the degradation of wild-type mRNAs. The participation of the NMD factors in general mRNA degradation suggests that they may regulate an aspect of translation termination common to all transcripts.
Figures

Similar articles
-
Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway.Mol Cell Biol. 1997 Mar;17(3):1580-94. doi: 10.1128/MCB.17.3.1580. Mol Cell Biol. 1997. PMID: 9032286 Free PMC article.
-
Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p.Mol Cell Biol. 2000 Jul;20(13):4591-603. doi: 10.1128/MCB.20.13.4591-4603.2000. Mol Cell Biol. 2000. PMID: 10848586 Free PMC article.
-
Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast.RNA. 1996 Feb;2(2):153-70. RNA. 1996. PMID: 8601282 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
Cited by
-
Analysis of the products of mRNA decapping and 3'-to-5' decay by denaturing gel electrophoresis.RNA. 2002 Jul;8(7):959-65. doi: 10.1017/s1355838202025049. RNA. 2002. PMID: 12166650 Free PMC article.
-
Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae.RNA. 2004 Apr;10(4):691-703. doi: 10.1261/rna.5147804. RNA. 2004. PMID: 15037778 Free PMC article.
-
Identification of a quality-control mechanism for mRNA 5'-end capping.Nature. 2010 Sep 30;467(7315):608-11. doi: 10.1038/nature09338. Epub 2010 Aug 29. Nature. 2010. PMID: 20802481 Free PMC article.
-
Decapping factor Dcp2 controls mRNA abundance and translation to adjust metabolism and filamentation to nutrient availability.Elife. 2023 Jun 2;12:e85545. doi: 10.7554/eLife.85545. Elife. 2023. PMID: 37266577 Free PMC article.
-
Ribosome-bound Upf1 forms distinct 80S complexes and conducts mRNA surveillance.RNA. 2022 Dec;28(12):1621-1642. doi: 10.1261/rna.079416.122. Epub 2022 Oct 3. RNA. 2022. PMID: 36192133 Free PMC article.
References
-
- Altamura N, Groudinsky O, Dujardin G, Slonimski P P. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. - PubMed
-
- Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. - PubMed
-
- Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
