Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA
- PMID: 11238851
- PMCID: PMC114118
- DOI: 10.1128/JVI.75.7.3250-3258.2001
Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA
Abstract
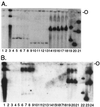
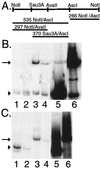
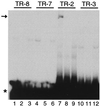
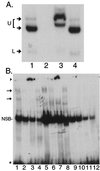
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) (also known as human herpesvirus 8) latently infects KS tumors, primary effusion lymphomas (PELs), and PEL cell lines. In latently infected cells, KSHV DNA is maintained as circularized, extrachromosomal episomes. To persist in proliferating cells, KSHV episomes must replicate and efficiently segregate to progeny nuclei. In uninfected B-lymphoblastoid cells, KSHV latency-associated nuclear antigen (LANA1) is necessary and sufficient for persistence of artificial episomes containing specific KSHV DNA. In previous work, the cis-acting sequence required for episome persistence contained KSHV terminal-repeat (TR) DNA and unique KSHV sequence. We now show that cis-acting KSHV TR DNA is necessary and sufficient for LANA1-mediated episome persistence. Furthermore, LANA1 binds TR DNA in mobility shift assays and a 20-nucleotide LANA1 binding sequence has been identified. Since LANA1 colocalizes with KSHV episomes along metaphase chromosomes, these results are consistent with a model in which LANA1 may bridge TR DNA to chromosomes during mitosis to efficiently segregate KSHV episomes to progeny nuclei.
Figures



Similar articles
-
The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence.J Virol. 2004 Jan;78(1):294-301. doi: 10.1128/jvi.78.1.294-301.2004. J Virol. 2004. PMID: 14671111 Free PMC article.
-
KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence.Virology. 2004 Feb 20;319(2):225-36. doi: 10.1016/j.virol.2003.11.002. Virology. 2004. PMID: 14980483
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
The KSHV latency-associated nuclear antigen: a multifunctional protein.Front Biosci. 2002 Mar 1;7:d726-30. doi: 10.2741/komatsu. Front Biosci. 2002. PMID: 11861213 Review.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats.J Virol. 2012 Sep;86(18):9983-94. doi: 10.1128/JVI.00839-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761383 Free PMC article.
-
Kaposi's sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):16287-92. doi: 10.1073/pnas.0703508104. Epub 2007 Oct 1. Proc Natl Acad Sci U S A. 2007. PMID: 17909182 Free PMC article.
-
ORF73 of herpesvirus saimiri, a viral homolog of Kaposi's sarcoma-associated herpesvirus, modulates the two cellular tumor suppressor proteins p53 and pRb.J Virol. 2004 Oct;78(19):10336-47. doi: 10.1128/JVI.78.19.10336-10347.2004. J Virol. 2004. PMID: 15367600 Free PMC article.
-
Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.J Virol. 2005 Nov;79(21):13618-29. doi: 10.1128/JVI.79.21.13618-13629.2005. J Virol. 2005. PMID: 16227282 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing.J Virol. 2007 Aug;81(15):8225-35. doi: 10.1128/JVI.00411-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522213 Free PMC article.
References
-
- Ausubel F M. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998.
-
- Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. - PubMed
-
- Bastien N, McBride A A. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology. 2000;270:124–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

