Role for human immunodeficiency virus type 1 Tat protein in suppression of viral reverse transcriptase activity during late stages of viral replication
- PMID: 11222691
- PMCID: PMC115892
- DOI: 10.1128/JVI.75.6.2675-2683.2001
Role for human immunodeficiency virus type 1 Tat protein in suppression of viral reverse transcriptase activity during late stages of viral replication
Abstract
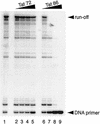
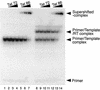
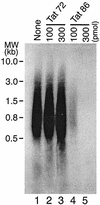
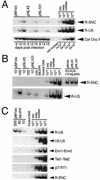
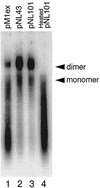
We have examined the role of the human immunodeficiency virus type 1 (HIV-1) Tat protein in the regulation of reverse transcription. We show that a two-exon but not a one-exon form of Tat markedly suppressed cell-free reverse transcriptase (RT) activity. Conversely, viruses expressing two-exon Tat (pNL43 and pNL101) showed rapid replication kinetics and more efficient endogenous RT activity compared with viruses expressing one-exon Tat (pM1ex). The pM1ex virions, as well as pM1ex-infected cells, also contained higher levels of viral DNA than did either the pNL43 or pNL101 viruses, indicating that reverse transcription might have continued during later stages of viral replication in the absence of the second Tat exon. Moreover, degradation of viral genomic RNA was more apparent in the pM1ex virions. Accordingly, we propose that the two-exon Tat may help augment viral infectivity by suppressing the reverse transcription reaction during late stages of viral synthesis and by preventing the synthesis of potentially deleterious viral DNA products.
Figures
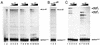
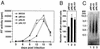
Similar articles
-
Suppression of an intrinsic strand transfer activity of HIV-1 Tat protein by its second-exon sequences.Virology. 2003 Mar 1;307(1):154-63. doi: 10.1016/s0042-6822(02)00068-5. Virology. 2003. PMID: 12667823
-
The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription.J Virol. 2002 Apr;76(8):3637-45. doi: 10.1128/jvi.76.8.3637-3645.2002. J Virol. 2002. PMID: 11907203 Free PMC article.
-
Tat is required for efficient HIV-1 reverse transcription.EMBO J. 1997 Mar 17;16(6):1224-35. doi: 10.1093/emboj/16.6.1224. EMBO J. 1997. PMID: 9135139 Free PMC article.
-
The role of Tat in HIV-1 replication: an activator and/or a suppressor?AIDS Rev. 2002 Jan-Mar;4(1):41-9. AIDS Rev. 2002. PMID: 11998784 Review.
-
RNA recognition and regulation of HIV-1 gene expression by viral factor Tat.Biochemistry (Mosc). 1998 May;63(5):489-503. Biochemistry (Mosc). 1998. PMID: 9632883 Review.
Cited by
-
PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA.Retrovirology. 2006 Dec 18;3:93. doi: 10.1186/1742-4690-3-93. Retrovirology. 2006. PMID: 17176473 Free PMC article.
-
Replication of human immunodeficiency viruses engineered with heterologous Tat-transactivation response element interactions.J Virol. 2003 Feb;77(3):1984-91. doi: 10.1128/jvi.77.3.1984-1991.2003. J Virol. 2003. PMID: 12525632 Free PMC article.
-
Specific Interaction between eEF1A and HIV RT Is Critical for HIV-1 Reverse Transcription and a Potential Anti-HIV Target.PLoS Pathog. 2015 Dec 1;11(12):e1005289. doi: 10.1371/journal.ppat.1005289. eCollection 2015 Dec. PLoS Pathog. 2015. PMID: 26624286 Free PMC article.
-
A mutant tat protein inhibits HIV-1 reverse transcription by targeting the reverse transcription complex.J Virol. 2015 May;89(9):4827-36. doi: 10.1128/JVI.03440-14. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673710 Free PMC article.
-
Y44A Mutation in the Acidic Domain of HIV-2 Tat Impairs Viral Reverse Transcription and LTR-Transactivation.Int J Mol Sci. 2020 Aug 17;21(16):5907. doi: 10.3390/ijms21165907. Int J Mol Sci. 2020. PMID: 32824587 Free PMC article.
References
-
- Arts E J, Li X, Gu Z, Kleiman L, Parniak M A, Wainberg M A. Comparison of deoxyoligonucleotide and tRNALys-3 as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J Biol Chem. 1994;269:14672–14680. - PubMed
-
- Biswas D K, Tius M A, Zhuo J, Pardee A B. Canventol inhibits HIV-1 replication by Tat-induced Tar-independent mechanism. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:120–127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

