The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity
- PMID: 11191109
- PMCID: PMC1507985
- DOI: 10.1038/sj.neo.7900112
The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity
Abstract
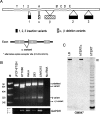
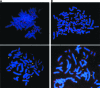
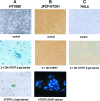
The telomerase catalytic subunit (hTERT) is an essential component of the holoenzyme complex that adds telomeric repeats to the ends of human chromosomes. Maintenance of telomeres by telomerase or another mechanism is required for cell immortalization, and loss of telomeric DNA has been proposed as a trigger for cellular senescence. Available evidence suggests that regulation of telomerase activity primarily depends on transcriptional control of hTERT. However, several human tissues as well as some normal cell strains have been shown to express low levels of hTERT mRNA even though they lack telomerase activity. We have previously identified six splice variants of hTERT, including a "deletion" variant (hTERTalpha) that is missing conserved residues from the catalytic core of the protein. Several of the deletion variants have been detected in normal and developing human tissues. We now show that hTERTalpha inhibits endogenous telomerase activity, which results in telomere shortening and chromosome end-to-end fusions. Telomerase inhibition induced a senescence-like state in HT1080 cells and apoptosis in a jejunal fibroblast cell line. These results suggest a possible role for hTERT splice variants in the regulation of telomerase activity.
Figures

Similar articles
-
An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity.Neoplasia. 2000 Sep-Oct;2(5):433-40. doi: 10.1038/sj.neo.7900113. Neoplasia. 2000. PMID: 11191110 Free PMC article.
-
Telomere dynamics, end-to-end fusions and telomerase activation during the human fibroblast immortalization process.Oncogene. 1999 Jul 22;18(29):4211-23. doi: 10.1038/sj.onc.1202797. Oncogene. 1999. PMID: 10435634
-
Comparison of human mammary epithelial cells immortalized by simian virus 40 T-Antigen or by the telomerase catalytic subunit.Oncogene. 2002 Jan 3;21(1):128-39. doi: 10.1038/sj.onc.1205014. Oncogene. 2002. PMID: 11791183
-
Telomeres, telomerase, and myc. An update.Mutat Res. 2000 Jan;462(1):31-47. doi: 10.1016/s1383-5742(99)00091-5. Mutat Res. 2000. PMID: 10648922 Review.
-
Telomere length maintenance in aging and carcinogenesis.Int J Oncol. 2000 Nov;17(5):981-9. doi: 10.3892/ijo.17.5.981. Int J Oncol. 2000. PMID: 11029502 Review.
Cited by
-
Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer.Nat Genet. 2013 Apr;45(4):371-84, 384e1-2. doi: 10.1038/ng.2566. Nat Genet. 2013. PMID: 23535731 Free PMC article.
-
Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene.Neoplasia. 2003 May-Jun;5(3):193-7. doi: 10.1016/S1476-5586(03)80051-9. Neoplasia. 2003. PMID: 12869302 Free PMC article.
-
Mechanism of telomerase activation by v-Rel and its contribution to transformation.J Virol. 2006 Jan;80(1):281-95. doi: 10.1128/JVI.80.1.281-295.2006. J Virol. 2006. PMID: 16352553 Free PMC article.
-
The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis.Cancer Res. 2013 May 1;73(9):2817-28. doi: 10.1158/0008-5472.CAN-12-3082. Epub 2013 Apr 22. Cancer Res. 2013. PMID: 23610451 Free PMC article.
-
Quantification of alternative splicing variants of human telomerase reverse transcriptase and correlations with telomerase activity in lung cancer.PLoS One. 2012;7(6):e38868. doi: 10.1371/journal.pone.0038868. Epub 2012 Jun 18. PLoS One. 2012. PMID: 22723897 Free PMC article.
References
-
- Colgin LM, Reddel RR. Telomere maintenance mechanisms and cellular immortalization. Curr Opin Genet Dev. 1999;9:97–103. - PubMed
-
- Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides. Dokl Akad Nauk SSR. 1971;201:1496–1499. - PubMed
-
- Harley CB. Telomere loss: Mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. - PubMed
-
- Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. - PubMed
-
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Amgen EST Program. Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997;275:973–977. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
