FTZ-Factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs
- PMID: 11157757
- PMCID: PMC133472
- DOI: 10.1093/emboj/20.3.510
FTZ-Factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs
Abstract
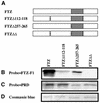
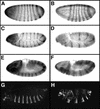
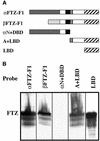
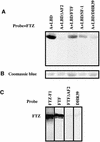
To activate transcription, most nuclear receptor proteins require coactivators that bind to their ligand-binding domains (LBDs). The Drosophila FTZ-Factor1 (FTZ-F1) protein is a conserved member of the nuclear receptor superfamily, but was previously thought to lack an AF2 motif, a motif that is required for ligand and coactivator binding. Here we show that FTZ-F1 does have an AF2 motif and that it is required to bind a coactivator, the homeodomain-containing protein Fushi tarazu (FTZ). We also show that FTZ contains an AF2-interacting nuclear receptor box, the first to be found in a homeodomain protein. Both interaction motifs are shown to be necessary for physical interactions in vitro and for functional interactions in developing embryos. These unexpected findings have important implications for the conserved homologs of the two proteins.
Figures

Similar articles
-
The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains.Mech Dev. 2001 Sep;107(1-2):39-53. doi: 10.1016/s0925-4773(01)00448-8. Mech Dev. 2001. PMID: 11520662
-
The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz.Nature. 1997 Feb 6;385(6616):552-5. doi: 10.1038/385552a0. Nature. 1997. PMID: 9020364
-
Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene.Development. 1997 Feb;124(4):839-47. doi: 10.1242/dev.124.4.839. Development. 1997. PMID: 9043065
-
Human Ad4BP/SF-1 and its related nuclear receptor.J Steroid Biochem Mol Biol. 1999 Apr-Jun;69(1-6):323-8. doi: 10.1016/s0960-0760(99)00081-3. J Steroid Biochem Mol Biol. 1999. PMID: 10419009 Review.
-
Orphan nuclear receptors adopted by crystallography.Curr Opin Struct Biol. 2005 Dec;15(6):708-15. doi: 10.1016/j.sbi.2005.10.009. Epub 2005 Nov 2. Curr Opin Struct Biol. 2005. PMID: 16263271 Review.
Cited by
-
Functional conservation of interactions between a homeodomain cofactor and a mammalian FTZ-F1 homologue.EMBO Rep. 2004 Jun;5(6):613-9. doi: 10.1038/sj.embor.7400147. Epub 2004 May 14. EMBO Rep. 2004. PMID: 15143342 Free PMC article.
-
Variation and constraint in Hox gene evolution.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):2211-6. doi: 10.1073/pnas.1210847110. Epub 2013 Jan 22. Proc Natl Acad Sci U S A. 2013. PMID: 23341600 Free PMC article.
-
LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1.Mol Cell Biol. 2003 Jan;23(1):238-49. doi: 10.1128/MCB.23.1.238-249.2003. Mol Cell Biol. 2003. PMID: 12482977 Free PMC article.
-
Interaction patterns of methoprene-tolerant and germ cell-expressed Drosophila JH receptors suggest significant differences in their functioning.Front Mol Biosci. 2023 Aug 15;10:1215550. doi: 10.3389/fmolb.2023.1215550. eCollection 2023. Front Mol Biosci. 2023. PMID: 37654797 Free PMC article.
-
Orphan nuclear receptor ftz-f1 (NR5A3) promotes egg chamber survival in the Drosophila ovary.G3 (Bethesda). 2021 Feb 9;11(2):jkab003. doi: 10.1093/g3journal/jkab003. G3 (Bethesda). 2021. PMID: 33693603 Free PMC article.
References
-
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. - PubMed
-
- Ayer S., Walker,N., Mosammaparast,M., Nelson,J.P., Shilo,B.Z. and Benyajati,C. (1993) Activation and repression of Drosophila alcohol dehydrogenase distal transcription by two steroid hormone receptor superfamily members binding to a common response element. Nucleic Acids Res., 21, 1619–1627. - PMC - PubMed
-
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. - PubMed
-
- Brook W.J. and Cohen,S.M. (1996) Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila leg. Science, 273, 1373–1377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

