Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion
- PMID: 11134073
- PMCID: PMC2150668
- DOI: 10.1083/jcb.151.7.1435
Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion
Abstract
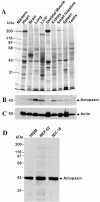
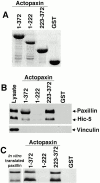
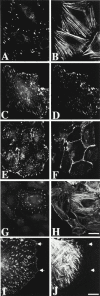
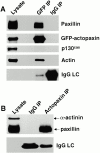
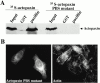
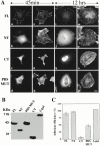
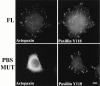
Paxillin is a focal adhesion adapter protein involved in the integration of growth factor- and adhesion-mediated signal transduction pathways. Paxillin LD motifs have been demonstrated to bind to several proteins associated with remodeling of the actin cytoskeleton including the focal adhesion kinase, vinculin, and a complex of proteins comprising p95PKL, PIX, and PAK (Turner, C.E., M. C. Brown, J.A. Perrotta, M.C. Riedy, S.N. Nikolopoulos, A.R. McDonald, S. Bagrodia, S. Thomas, and P.S. Leventhal. 1999. J. Cell Biol. 145:851-863). In this study, we report the cloning and initial characterization of a new paxillin LD motif-binding protein, actopaxin. Analysis of the deduced amino acid sequence of actopaxin reveals a 42-kD protein with two calponin homology domains and a paxillin-binding subdomain (PBS). Western blotting identifies actopaxin as a widely expressed protein. Actopaxin binds directly to both F-actin and paxillin LD1 and LD4 motifs. It exhibits robust focal adhesion localization in several cultured cell types but is not found along the length of the associated actin-rich stress fibers. Similar to paxillin, it is absent from actin-rich cell-cell adherens junctions. Also, actopaxin colocalizes with paxillin to rudimentary focal complexes at the leading edge of migrating cells. An actopaxin PBS mutant incapable of binding paxillin in vitro cannot target to focal adhesions when expressed in fibroblasts. In addition, ectopic expression of the PBS mutant and/or the COOH terminus of actopaxin in HeLa cells resulted in substantial reduction in adhesion to collagen. Together, these results suggest an important role for actopaxin in integrin-dependent remodeling of the actin cytoskeleton during cell motility and cell adhesion.
Figures



Similar articles
-
Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: A role in cytoskeletal remodeling.J Cell Biol. 1999 May 17;145(4):851-63. doi: 10.1083/jcb.145.4.851. J Cell Biol. 1999. PMID: 10330411 Free PMC article.
-
Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions.J Biol Chem. 2001 Jun 29;276(26):23499-505. doi: 10.1074/jbc.M102163200. Epub 2001 Apr 13. J Biol Chem. 2001. PMID: 11304546
-
Molecular dissection of actopaxin-integrin-linked kinase-Paxillin interactions and their role in subcellular localization.J Biol Chem. 2002 Jan 11;277(2):1568-75. doi: 10.1074/jbc.M108612200. Epub 2001 Nov 1. J Biol Chem. 2002. PMID: 11694518
-
Paxillin interactions.J Cell Sci. 2000 Dec;113 Pt 23:4139-40. doi: 10.1242/jcs.113.23.4139. J Cell Sci. 2000. PMID: 11069756 Review.
-
The paxillin LD motifs.FEBS Lett. 2002 Feb 20;513(1):114-8. doi: 10.1016/s0014-5793(01)03244-6. FEBS Lett. 2002. PMID: 11911889 Review.
Cited by
-
Rsu1 contributes to regulation of cell adhesion and spreading by PINCH1-dependent and - independent mechanisms.J Cell Commun Signal. 2013 Dec;7(4):279-93. doi: 10.1007/s12079-013-0207-5. Epub 2013 Jun 14. J Cell Commun Signal. 2013. PMID: 23765260 Free PMC article.
-
Targeted deletion of integrin-linked kinase reveals a role in T-cell chemotaxis and survival.Mol Cell Biol. 2005 Dec;25(24):11145-55. doi: 10.1128/MCB.25.24.11145-11155.2005. Mol Cell Biol. 2005. PMID: 16314534 Free PMC article.
-
TRIM15 is a focal adhesion protein that regulates focal adhesion disassembly.J Cell Sci. 2014 Sep 15;127(Pt 18):3928-42. doi: 10.1242/jcs.143537. Epub 2014 Jul 11. J Cell Sci. 2014. PMID: 25015296 Free PMC article.
-
Meta-fibrosis links positive energy balance and mitochondrial metabolism to insulin resistance.F1000Res. 2017 Sep 27;6:1758. doi: 10.12688/f1000research.11653.1. eCollection 2017. F1000Res. 2017. PMID: 29043068 Free PMC article. Review.
-
α-Parvin Expression in Breast Cancer Tissues: Correlation with Clinical Parameters and Prognostic Significance.Cells. 2024 Sep 19;13(18):1572. doi: 10.3390/cells13181572. Cells. 2024. PMID: 39329755 Free PMC article.
References
-
- Bagrodia S., Cerione R.A. PAK to the future. Trends Cell Biol. 1999;9:350–355. - PubMed
-
- Bagrodia S., Taylor S.J., Jordon K.A., Van Aelst L., Cerione R.A. A novel regulator of p21-activated kinases. J. Biol. Chem. 1998;273:23633–23636. - PubMed
-
- Banuelos S., Saraste M., Carugo K.D. Structural comparisons of calponin homology domainsimplications for actin binding. Structure. 1998;6:1419–1431. - PubMed
-
- Brown M.C., Curtis M.S., Turner C.E. Paxillin LD motifs may define a new family of protein recognition domains Nat. Struct. Biol 5 1998. 677 678a - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

