spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily
- PMID: 11114162
- PMCID: PMC18952
- DOI: 10.1073/pnas.011446498
spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily
Abstract
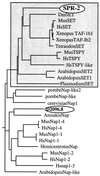
Presenilin plays critical roles in the genesis of Alzheimer's disease and in LIN-12/Notch signaling during development. Here, we describe a screen for genes that influence presenilin level or activity in Caenorhabditis elegans. We identified four spr (suppressor of presenilin) genes by reverting the egg-laying defective phenotype caused by a null allele of the sel-12 presenilin gene. We analyzed the spr-2 gene in some detail. We show that loss of spr-2 activity suppresses the egg-laying defective phenotype of different sel-12 alleles and requires activity of the hop-1 presenilin gene, suggesting that suppression is accomplished by elevating presenilin activity rather than by bypassing the need for presenilin activity. We also show that SPR-2 is a nuclear protein and is a member of a protein subfamily that includes human SET, which has been identified in numerous different biochemical assays and at translocation breakpoints associated with a subtype of acute myeloid leukemia.
Figures

Similar articles
-
Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans.Genes Dev. 2002 Oct 15;16(20):2713-28. doi: 10.1101/gad.1022402. Genes Dev. 2002. PMID: 12381669 Free PMC article.
-
Two suppressors of sel-12 encode C2H2 zinc-finger proteins that regulate presenilin transcription in Caenorhabditis elegans.Development. 2003 May;130(10):2117-28. doi: 10.1242/dev.00429. Development. 2003. PMID: 12668626
-
HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling.Proc Natl Acad Sci U S A. 1997 Oct 28;94(22):12204-9. doi: 10.1073/pnas.94.22.12204. Proc Natl Acad Sci U S A. 1997. PMID: 9342387 Free PMC article.
-
Loss of spr-5 bypasses the requirement for the C.elegans presenilin sel-12 by derepressing hop-1.EMBO J. 2002 Nov 1;21(21):5787-96. doi: 10.1093/emboj/cdf561. EMBO J. 2002. PMID: 12411496 Free PMC article.
-
Notch and presenilin: a proteolytic mechanism emerges.Curr Opin Cell Biol. 2001 Oct;13(5):627-34. doi: 10.1016/s0955-0674(00)00261-1. Curr Opin Cell Biol. 2001. PMID: 11544033 Review.
Cited by
-
Finding minimum gene subsets with heuristic breadth-first search algorithm for robust tumor classification.BMC Bioinformatics. 2012 Jul 25;13:178. doi: 10.1186/1471-2105-13-178. BMC Bioinformatics. 2012. PMID: 22830977 Free PMC article.
-
SPR-1/CoREST facilitates the maternal epigenetic reprogramming of the histone demethylase SPR-5/LSD1.Genetics. 2023 Mar 2;223(3):iyad005. doi: 10.1093/genetics/iyad005. Genetics. 2023. PMID: 36655746 Free PMC article.
-
Modeling Alzheimer's disease: from past to future.Front Pharmacol. 2013 Jun 19;4:77. doi: 10.3389/fphar.2013.00077. eCollection 2013. Front Pharmacol. 2013. PMID: 23801962 Free PMC article.
-
Mutations in genes involved in nonsense mediated decay ameliorate the phenotype of sel-12 mutants with amber stop mutations in Caenorhabditis elegans.BMC Genet. 2009 Mar 20;10:14. doi: 10.1186/1471-2156-10-14. BMC Genet. 2009. PMID: 19302704 Free PMC article.
-
Understanding the molecular basis of Alzheimer's disease using a Caenorhabditis elegans model system.Brain Struct Funct. 2010 Mar;214(2-3):263-83. doi: 10.1007/s00429-009-0235-3. Epub 2009 Dec 11. Brain Struct Funct. 2010. PMID: 20012092 Free PMC article. Review.
References
-
- Selkoe D J. Trends Cell Biol. 1998;8:447–453. - PubMed
-
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. - PubMed
-
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Nat Cell Biol. 2000;2:461–462. - PubMed
-
- Steiner H, Duff K, Capell A, Romig H, Grim M G, Lincoln S, Hardy J, Yu X, Picciano M, Fechteler K, et al. J Biol Chem. 1999;274:28669–28673. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

