The solution structure of the C-terminal domain of the Mu B transposition protein
- PMID: 11060014
- PMCID: PMC305798
- DOI: 10.1093/emboj/19.21.5625
The solution structure of the C-terminal domain of the Mu B transposition protein
Abstract
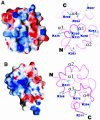
Mu B is one of four proteins required for the strand transfer step of bacteriophage Mu DNA transposition and the only one where no high resolution structural data is available. Structural work on Mu B has been hampered primarily by solubility problems and its tendency to aggregate. We have overcome this problem by determination of the three-dimensional structure of the C-terminal domain of Mu B (B(223-312)) in 1.5 M NaCl using NMR spectroscopic methods. The structure of Mu B(223-312) comprises four helices (backbone r.m.s.d. 0.46 A) arranged in a loosely packed bundle and resembles that of the N-terminal region of the replication helicase, DnaB. This structural motif is likely to be involved in the inter-domainal regulation of ATPase activity for both Mu A and DnaB. The approach described here for structural determination in high salt may be generally applicable for proteins that do not crystallize and that are plagued by solubility problems at low ionic strength.
Figures




Similar articles
-
Mechanism of DNA binding by the DnaB helicase of Escherichia coli: analysis of the roles of domain gamma in DNA binding.Biochemistry. 1999 Aug 24;38(34):10929-39. doi: 10.1021/bi990049l. Biochemistry. 1999. PMID: 10460148
-
Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate.J Mol Biol. 1999 Jun 11;289(3):503-16. doi: 10.1006/jmbi.1999.2783. J Mol Biol. 1999. PMID: 10356325
-
NMR structure and functional studies of the Mu repressor DNA-binding domain.Biochemistry. 1999 Jun 29;38(26):8367-76. doi: 10.1021/bi990530b. Biochemistry. 1999. PMID: 10387082
-
Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding, and oligomerization.Biochemistry. 1999 Aug 24;38(34):10919-28. doi: 10.1021/bi990048t. Biochemistry. 1999. PMID: 10460147
-
Conformational dynamics of DnaB helicase upon DNA and nucleotide binding: analysis by intrinsic tryptophan fluorescence quenching.Biochemistry. 2003 Feb 25;42(7):1910-21. doi: 10.1021/bi025992v. Biochemistry. 2003. PMID: 12590577
Cited by
-
MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition.Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):E2441-50. doi: 10.1073/pnas.1309499110. Epub 2013 Jun 17. Proc Natl Acad Sci U S A. 2013. PMID: 23776210 Free PMC article.
-
Dynamics of a protein polymer: the assembly and disassembly pathways of the MuB transposition target complex.EMBO J. 2002 Mar 15;21(6):1477-86. doi: 10.1093/emboj/21.6.1477. EMBO J. 2002. PMID: 11889053 Free PMC article.
-
MuB gives a new twist to target DNA selection.Mob Genet Elements. 2013 Sep 1;3(5):e27515. doi: 10.4161/mge.27515. Epub 2013 Dec 12. Mob Genet Elements. 2013. PMID: 24478936 Free PMC article.
-
Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.Nat Commun. 2024 Jul 31;15(1):6445. doi: 10.1038/s41467-024-50722-1. Nat Commun. 2024. PMID: 39085263 Free PMC article.
-
Transposable Phage Mu.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.MDNA3-0007-2014. doi: 10.1128/microbiolspec.MDNA3-0007-2014. Microbiol Spectr. 2014. PMID: 26104374 Free PMC article. Review.
References
-
- Adzuma K. and Mizuuchi,K. (1991) Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage mu. Involvement of protein oligomerization in the ATPase cycle. J. Biol. Chem., 266, 6159–6167. - PubMed
-
- Baker T.A., Mizuuchi,M. and Mizuuchi,K. (1991) MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell, 65, 1003–1013. - PubMed
-
- Biswas S.B., Chen,P.H. and Biswas,E.E. (1994) Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry, 33, 11307–11314. - PubMed
-
- Brunger A.T. (1992) X-PLOR version 3.1: A system for X-ray crystallography and NMR. Yale University, New Haven, CT.
-
- Chaconas G. (1987) Phage Mu. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

