Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis
- PMID: 11053391
- PMCID: PMC94793
- DOI: 10.1128/JB.182.22.6456-6462.2000
Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis
Abstract
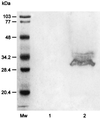
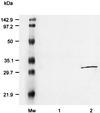
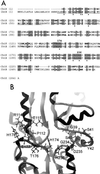
Porphyromonas gingivalis is a gram-negative, anaerobic coccobacillus that has been implicated as a major etiological agent in the development of chronic periodontitis. In this paper, we report the characterization of a protein, IhtB (iron heme transport; formerly designated Pga30), that is an outer membrane hemin-binding protein potentially involved in iron assimilation by P. gingivalis. IhtB was localized to the cell surface of P. gingivalis by Western blot analysis of a Sarkosyl-insoluble outer membrane preparation and by immunocytochemical staining of whole cells using IhtB peptide-specific antisera. The protein, released from the cell surface, was shown to bind to hemin using hemin-agarose. The growth of heme-limited, but not heme-replete, P. gingivalis cells was inhibited by preincubation with IhtB peptide-specific antisera. The ihtB gene was located between an open reading frame encoding a putative TonB-linked outer membrane receptor and three open reading frames that have sequence similarity to ATP binding cassette transport system operons in other bacteria. Analysis of the deduced amino acid sequence of IhtB showed significant similarity to the Salmonella typhimurium protein CbiK, a cobalt chelatase that is structurally related to the ATP-independent family of ferrochelatases. Molecular modeling indicated that the IhtB amino acid sequence could be threaded onto the CbiK fold with the IhtB structural model containing the active-site residues critical for chelatase activity. These results suggest that IhtB is a peripheral outer membrane chelatase that may remove iron from heme prior to uptake by P. gingivalis.
Figures


Similar articles
-
The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin.J Bacteriol. 1997 Aug;179(15):4778-88. doi: 10.1128/jb.179.15.4778-4788.1997. J Bacteriol. 1997. PMID: 9244265 Free PMC article.
-
Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis.J Bacteriol. 2000 Oct;182(20):5737-48. doi: 10.1128/JB.182.20.5737-5748.2000. J Bacteriol. 2000. PMID: 11004172 Free PMC article.
-
Characterization and expression of a novel Porphyromonas gingivalis outer membrane protein, Omp28.Oral Microbiol Immunol. 2002 Jun;17(3):150-6. doi: 10.1034/j.1399-302x.2002.170303.x. Oral Microbiol Immunol. 2002. PMID: 12030966
-
Regulation of hemin and iron transport in Porphyromonas gingivalis.Adv Dent Res. 1995 Feb;9(1):41-7. doi: 10.1177/08959374950090010801. Adv Dent Res. 1995. PMID: 7669213 Review.
-
Iron and heme utilization in Porphyromonas gingivalis.FEMS Microbiol Rev. 2005 Jan;29(1):119-44. doi: 10.1016/j.femsre.2004.09.001. FEMS Microbiol Rev. 2005. PMID: 15652979 Review.
Cited by
-
LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis.Infect Immun. 2006 Jul;74(7):3834-44. doi: 10.1128/IAI.01768-05. Infect Immun. 2006. PMID: 16790755 Free PMC article.
-
Outer membrane lipoprotein biogenesis: Lol is not the end.Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150030. doi: 10.1098/rstb.2015.0030. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370942 Free PMC article. Review.
-
Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins.J Bacteriol. 2001 Oct;183(19):5599-608. doi: 10.1128/JB.183.19.5599-5608.2001. J Bacteriol. 2001. PMID: 11544222 Free PMC article.
-
Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis.J Bacteriol. 2001 Oct;183(19):5609-16. doi: 10.1128/JB.183.19.5609-5616.2001. J Bacteriol. 2001. PMID: 11544223 Free PMC article.
-
Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti.Appl Environ Microbiol. 2002 Dec;68(12):5877-81. doi: 10.1128/AEM.68.12.5877-5881.2002. Appl Environ Microbiol. 2002. PMID: 12450806 Free PMC article.
References
-
- Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure. 1997;5:1501–1510. - PubMed
-
- Beall B, Sanden G N. A Bordetella pertussis FepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. - PubMed
-
- Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. - PubMed
-
- Bramanti T, Holt S. Iron-regulated outer membrane proteins in the periodontopathogenic bacterium Bacteroides gingivalis. Biochem Biophys Res Commun. 1990;166:1146–1154. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

