DNA replication progresses on the periphery of nuclear aggregates formed by the BCL6 transcription factor
- PMID: 11046151
- PMCID: PMC102161
- DOI: 10.1128/MCB.20.22.8560-8570.2000
DNA replication progresses on the periphery of nuclear aggregates formed by the BCL6 transcription factor
Abstract
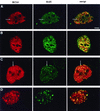
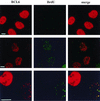
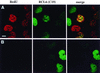
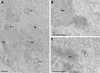
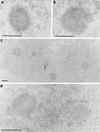
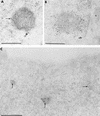
The BCL6 proto-oncogene, frequently alterated in non-Hodgkin lymphoma, encodes a POZ/zinc finger protein that localizes into discrete nuclear subdomains. Upon prolonged BCL6 overexpression in cells bearing an inducible BCL6 allele (UTA-L cells), these subdomains apparently coincide with sites of DNA synthesis. Here, we explore the relationship between BCL6 and replication by both electron and confocal laser scanning microscopy. First, by electron microscope analyses, we found that endogenous BCL6 is associated with replication foci. Moreover, we show that a relatively low expression level of BCL6 reached after a brief induction in UTA-L cells is sufficient to observe its targeting to mid, late, and at least certain early replication foci visualized by a pulse-labeling with bromodeoxyuridine (BrdU). In addition, when UTA-L cells are simultaneously induced for BCL6 expression and exposed to BrdU for a few hours just after the release from a block in mitosis, a nuclear diffuse BCL6 staining indicates cells in G(1), while cells in S show a more punctate nuclear BCL6 distribution associated with replication foci. Finally, ultrastructural analyses in UTA-L cells exposed to BrdU for various times reveal that replication progresses just around, but not within, BCL6 subdomains. Thus, nascent DNA is localized near, but not colocalized with, BCL6 subdomains, suggesting that they play an architectural role influencing positioning and/or assembly of replication foci. Together with its previously function as transcription repressor recruiting a histone deacetylase complex, BCL6 may therefore contribute to link nuclear organization, replication, and chromatin-mediated regulation.
Figures
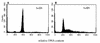
Similar articles
-
The relationship between BCL6 bodies and nuclear sites of normal and halogenated DNA and RNA synthesis.Microsc Res Tech. 2003 Jul 1;61(4):389-407. doi: 10.1002/jemt.10363. Microsc Res Tech. 2003. PMID: 12811744
-
Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF.Oncogene. 2000 Dec 14;19(54):6240-50. doi: 10.1038/sj.onc.1203976. Oncogene. 2000. PMID: 11175338
-
Overexpressed BCL6 (LAZ3) oncoprotein triggers apoptosis, delays S phase progression and associates with replication foci.Oncogene. 1999 Sep 9;18(36):5063-75. doi: 10.1038/sj.onc.1202892. Oncogene. 1999. PMID: 10490843
-
Targeted somatic mutation of the BCL6 proto-oncogene and its impact on lymphomagenesis.Hematology. 2005 Apr;10(2):115-29. doi: 10.1080/10245330400026105. Hematology. 2005. PMID: 16019457 Review.
-
Internal deletions within the BCL6 gene in B-cell non-Hodgkin's lymphoma.Leuk Lymphoma. 2000 Aug;38(5-6):505-12. doi: 10.3109/10428190009059269. Leuk Lymphoma. 2000. PMID: 10953971 Review.
Cited by
-
Inhibition of the transcriptional repressor complex Bcl-6/BCoR induces endothelial sprouting but does not promote tumor growth.Oncotarget. 2017 Jan 3;8(1):552-564. doi: 10.18632/oncotarget.13477. Oncotarget. 2017. PMID: 27880939 Free PMC article.
-
High-intensity UV laser ChIP-seq for the study of protein-DNA interactions in living cells.Nat Commun. 2017 Nov 3;8(1):1303. doi: 10.1038/s41467-017-01251-7. Nat Commun. 2017. PMID: 29101361 Free PMC article.
-
Overexpression of the transcriptional repressor complex BCL-6/BCoR leads to nuclear aggregates distinct from classical aggresomes.PLoS One. 2013 Oct 11;8(10):e76845. doi: 10.1371/journal.pone.0076845. eCollection 2013. PLoS One. 2013. PMID: 24146931 Free PMC article.
-
DNA Replication: From Radioisotopes to Click Chemistry.Molecules. 2018 Nov 17;23(11):3007. doi: 10.3390/molecules23113007. Molecules. 2018. PMID: 30453631 Free PMC article. Review.
References
-
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein/protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1996;6:1495–1503. - PubMed
-
- Albagli O, Dhordain P, Lantoine D, Auradé F, Quief S, Kerckaert J P, Montarras D, Pinset C. Increased expression of the LAZ3(BCL6) proto-oncogene accompanies murine skeletal myogenesis. Differentiation. 1998;64:33–44. - PubMed
-
- Albagli O, Lantoine D, Quief S, Quignon F, Englert C, Kerckaert J P, Montarras D, Pinset C, Lindon C. Overexpressed BCL6 (LAZ3) oncoprotein triggers apoptosis, delays S phase progression and associates with replication foci. Oncogene. 1999;18:5063–5075. - PubMed
-
- Almouzni G, Wolffe A P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
