Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg
- PMID: 11044080
- PMCID: PMC110910
- DOI: 10.1128/jvi.74.22.10359-10370.2000
Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg
Abstract
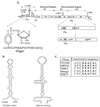
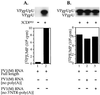
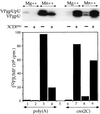
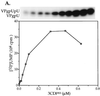
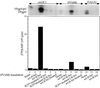
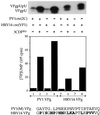
The first step in the replication of the plus-stranded poliovirus RNA is the synthesis of a complementary minus strand. This process is initiated by the covalent attachment of UMP to the terminal protein VPg, yielding VPgpU and VPgpUpU. We have previously shown that these products can be made in vitro in a reaction that requires only synthetic VPg, UTP, poly(A), purified poliovirus RNA polymerase 3D(pol), and Mg(2+) (A. V. Paul, J. H. van Boom, D. Filippov, and E. Wimmer, Nature 393:280-284, 1998). Since such a poly(A)-dependent process cannot confer sufficient specificity to poliovirus RNA replication, we have developed a new assay to search for a viral RNA template in conjunction with viral or cellular factors that could provide this function. We have now discovered a small RNA hairpin in the coding region of protein 2C as the site in PV1(M) RNA that is used as the primary template for the in vitro uridylylation of VPg. This hairpin has recently been described in poliovirus RNA as being an essential structure for the initiation of minus strand RNA synthesis (I. Goodfellow, Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans, J. Virol. 74:4590-4600, 2000). The uridylylation reaction either with transcripts of cre(2C) RNA or with full-length PV1(M) RNA as the template is strongly stimulated by the addition of purified viral protein 3CD(pro). Deletion of the cre(2C) RNA sequences from minigenomes eliminates their ability to serve as template in the reaction. A similar signal in the coding region of VP1 in HRV14 RNA (K. L. McKnight and S. M. Lemon, RNA 4:1569-1584, 1998) and the poliovirus cre(2C) can be functionally exchanged in the assay. The mechanism by which the VPgpUpU precursor, made specifically on the cre(2C) template, might be transferred to the site where it serves as primer for poliovirus RNA synthesis, remains to be determined.
Figures




Similar articles
-
Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation.J Virol. 2000 Nov;74(22):10371-80. doi: 10.1128/jvi.74.22.10371-10380.2000. J Virol. 2000. PMID: 11044081 Free PMC article.
-
Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro).J Virol. 2001 Nov;75(22):10979-90. doi: 10.1128/JVI.75.22.10979-10990.2001. J Virol. 2001. PMID: 11602738 Free PMC article.
-
Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA.J Virol. 2003 May;77(9):5152-66. doi: 10.1128/jvi.77.9.5152-5166.2003. J Virol. 2003. PMID: 12692218 Free PMC article.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
-
Uridylylation of the genome-linked protein of poliovirus in vitro is dependent upon an endogenous RNA template.Virus Res. 1987 Sep;8(3):193-204. doi: 10.1016/0168-1702(87)90015-3. Virus Res. 1987. PMID: 2825442 Review.
Cited by
-
Additive Promotion of Viral Internal Ribosome Entry Site-Mediated Translation by Far Upstream Element-Binding Protein 1 and an Enterovirus 71-Induced Cleavage Product.PLoS Pathog. 2016 Oct 25;12(10):e1005959. doi: 10.1371/journal.ppat.1005959. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27780225 Free PMC article.
-
The genome-linked protein VPg of vertebrate viruses - a multifaceted protein.Curr Opin Virol. 2011 Nov;1(5):355-62. doi: 10.1016/j.coviro.2011.09.003. Epub 2011 Oct 7. Curr Opin Virol. 2011. PMID: 22440837 Free PMC article. Review.
-
Enterovirus 71 VPg uridylation uses a two-molecular mechanism of 3D polymerase.J Virol. 2012 Dec;86(24):13662-71. doi: 10.1128/JVI.01712-12. Epub 2012 Oct 10. J Virol. 2012. PMID: 23055549 Free PMC article.
-
High-resolution structure of a picornaviral internal cis-acting RNA replication element (cre).Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12688-93. doi: 10.1073/pnas.0403079101. Epub 2004 Aug 16. Proc Natl Acad Sci U S A. 2004. PMID: 15314212 Free PMC article.
-
Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor.J Virol. 2005 Jun;79(11):7207-16. doi: 10.1128/JVI.79.11.7207-7216.2005. J Virol. 2005. PMID: 15890959 Free PMC article.
References
-
- Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. - PubMed
-
- Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–5266. - PubMed
-
- Andino R, Rieckhof E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
-
- Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–37069. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

