Wheat cytosolic acetyl-CoA carboxylase complements an ACC1 null mutation in yeast
- PMID: 11038571
- PMCID: PMC23321
- DOI: 10.1073/pnas.94.18.9990
Wheat cytosolic acetyl-CoA carboxylase complements an ACC1 null mutation in yeast
Abstract
Spores harboring an ACC1 deletion derived from a diploid Saccharomyces cerevisiae strain, in which one copy of the entire ACC1 gene is replaced with a LEU2 cassette, fail to grow. A chimeric gene consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat cytosolic acetyl-CoA carboxylase (ACCase) cDNA, and yeast ACC1 3' tail was used to complement a yeast ACC1 mutation. The complementation demonstrates that active wheat ACCase can be produced in yeast. At low concentrations of galactose, the activity of the "wheat gene" driven by the GAL10 promoter is low and ACCase becomes limiting for growth, a condition expected to enhance transgenic yeast sensitivity to wheat ACCase-specific inhibitors. An aryloxyphenoxypropionate and two cyclohexanediones do not inhibit growth of haploid yeast strains containing the yeast ACC1 gene, but one cyclohexanedione inhibits growth of the gene-replacement strains at concentrations below 0.2 mM. In vitro, the activity of wheat cytosolic ACCase produced by the gene-replacement yeast strain is inhibited by haloxyfop and cethoxydim at concentrations above 0.02 mM. The activity of yeast ACCase is less affected. The wheat plastid ACCase in wheat germ extract is inhibited by all three herbicides at concentrations below 0.02 mM. Yeast gene-replacement strains will provide a convenient system for the study of plant ACCases.
Figures
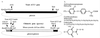

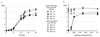
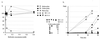

Similar articles
-
An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors.Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6617-22. doi: 10.1073/pnas.121172798. Epub 2001 May 29. Proc Natl Acad Sci U S A. 2001. PMID: 11381131 Free PMC article.
-
Herbicide sensitivity determinant of wheat plastid acetyl-CoA carboxylase is located in a 400-amino acid fragment of the carboxyltransferase domain.Proc Natl Acad Sci U S A. 1999 Dec 7;96(25):14647-51. doi: 10.1073/pnas.96.25.14647. Proc Natl Acad Sci U S A. 1999. PMID: 10588759 Free PMC article.
-
Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1.mBio. 2014 May 6;5(3):e01130-14. doi: 10.1128/mBio.01130-14. mBio. 2014. PMID: 24803522 Free PMC article.
-
Comprehensive guide to acetyl-carboxylases in algae.Crit Rev Biotechnol. 2013 Mar;33(1):49-65. doi: 10.3109/07388551.2012.668671. Epub 2012 Apr 23. Crit Rev Biotechnol. 2013. PMID: 22524446 Review.
-
Resistance to acetyl-CoA carboxylase-inhibiting herbicides.Pest Manag Sci. 2014 Sep;70(9):1405-17. doi: 10.1002/ps.3790. Epub 2014 May 6. Pest Manag Sci. 2014. PMID: 24700409 Review.
Cited by
-
Recombinant yeast screen for new inhibitors of human acetyl-CoA carboxylase 2 identifies potential drugs to treat obesity.Proc Natl Acad Sci U S A. 2010 May 18;107(20):9093-8. doi: 10.1073/pnas.1003721107. Epub 2010 May 3. Proc Natl Acad Sci U S A. 2010. PMID: 20439761 Free PMC article.
-
An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors.Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6617-22. doi: 10.1073/pnas.121172798. Epub 2001 May 29. Proc Natl Acad Sci U S A. 2001. PMID: 11381131 Free PMC article.
-
Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass.Plant Physiol. 2005 Mar;137(3):794-806. doi: 10.1104/pp.104.046144. Epub 2004 Dec 3. Plant Physiol. 2005. PMID: 15579665 Free PMC article.
-
A unified molecular mechanism for the regulation of acetyl-CoA carboxylase by phosphorylation.Cell Discov. 2016 Nov 29;2:16044. doi: 10.1038/celldisc.2016.44. eCollection 2016. Cell Discov. 2016. PMID: 27990296 Free PMC article.
-
Expression of Yarrowia lipolytica acetyl-CoA carboxylase in Saccharomyces cerevisiae and its effect on in-vivo accumulation of Malonyl-CoA.Comput Struct Biotechnol J. 2022 Jan 22;20:779-787. doi: 10.1016/j.csbj.2022.01.020. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36284710 Free PMC article.
References
-
- Konishi T, Shinohara K, Yamada K, Sasaki Y. Plant Cell Physiol. 1996;37:117–122. - PubMed
-
- Haslacher M, Ivessa A, Paltauf F, Kohlwein S. J Biol Chem. 1993;268:10946–10952. - PubMed
-
- Golz A, Focke M, Lichtenthaler H K. J Plant Physiol. 1994;143:426–433.
-
- Holt J S, Powles S B, Holtum J A M. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:203–229.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

