Functional selectivity of recombinant mammalian SWI/SNF subunits
- PMID: 11018012
- PMCID: PMC316972
- DOI: 10.1101/gad.828000
Functional selectivity of recombinant mammalian SWI/SNF subunits
Abstract
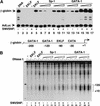
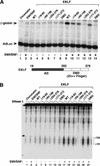
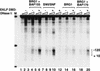
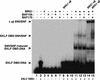
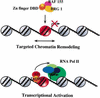
The SWI/SNF family of chromatin-remodeling complexes plays a key role in facilitating the binding of specific transcription factors to nucleosomal DNA in diverse organisms from yeast to man. Yet the process by which SWI/SNF and other chromatin-remodeling complexes activate specific subsets of genes is poorly understood. We show that mammalian SWI/SNF regulates transcription from chromatin-assembled genes in a factor-specific manner in vitro. The DNA-binding domains (DBDs) of several zinc finger proteins, including EKLF, interact directly with SWI/SNF to generate DNase I hypersensitivity within the chromatin-assembled beta-globin promoter. Interestingly, we find that two SWI/SNF subunits (BRG1 and BAF155) are necessary and sufficient for targeted chromatin remodeling and transcriptional activation by EKLF in vitro. Remodeling is achieved with only the BRG1-BAF155 minimal complex and the EKLF zinc finger DBD, whereas transcription requires, in addition, an activation domain. In contrast, the BRG1-BAF155 complex does not interact or function with two unrelated transcription factors, TFE3 and NF-kappaB. We conclude that specific domains of certain transcription factors differentially target SWI/SNF complexes to chromatin in a gene-selective manner and that individual SWI/SNF subunits play unique roles in transcription factor-directed nucleosome remodeling.
Figures
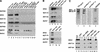

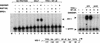
Similar articles
-
A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro.Cell. 1998 Oct 2;95(1):93-104. doi: 10.1016/s0092-8674(00)81785-7. Cell. 1998. PMID: 9778250
-
Distinct domains of erythroid Krüppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter.Mol Cell Biol. 2002 Jan;22(1):161-70. doi: 10.1128/MCB.22.1.161-170.2002. Mol Cell Biol. 2002. PMID: 11739731 Free PMC article.
-
Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article.
-
BAFfling pathologies: Alterations of BAF complexes in cancer.Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review.
-
Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence.Biochem Cell Biol. 2007 Aug;85(4):419-25. doi: 10.1139/O07-070. Biochem Cell Biol. 2007. PMID: 17713577 Review.
Cited by
-
SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes.PLoS One. 2012;7(3):e33834. doi: 10.1371/journal.pone.0033834. Epub 2012 Mar 19. PLoS One. 2012. PMID: 22442726 Free PMC article.
-
Advances in the understanding of haemoglobin switching.Br J Haematol. 2010 Apr;149(2):181-94. doi: 10.1111/j.1365-2141.2010.08105.x. Epub 2010 Mar 1. Br J Haematol. 2010. PMID: 20201948 Free PMC article. Review.
-
Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation.Mol Cell Biol. 2007 Jun;27(12):4551-65. doi: 10.1128/MCB.00235-07. Epub 2007 Apr 16. Mol Cell Biol. 2007. PMID: 17438135 Free PMC article.
-
Stage-specific repression by the EKLF transcriptional activator.Mol Cell Biol. 2004 Dec;24(23):10416-24. doi: 10.1128/MCB.24.23.10416-10424.2004. Mol Cell Biol. 2004. PMID: 15542849 Free PMC article.
-
Identification of a polymorphic, neuron-specific chromatin remodeling complex.Genes Dev. 2002 Oct 1;16(19):2509-17. doi: 10.1101/gad.992102. Genes Dev. 2002. PMID: 12368262 Free PMC article.
References
-
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
