Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase
- PMID: 10982868
- PMCID: PMC110723
- DOI: 10.1093/nar/28.18.3497
Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase
Abstract
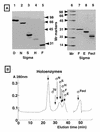
Seven different species of the RNA polymerase sigma subunit exist in Escherichia coli, each binding to a single species of the core enzyme and thereby directing transcription of a specific set of genes. To test the sigma competition model in the global regulation of gene transcription, all seven E.coli sigma subunits have been purified and compared for their binding affinities to the same core RNA polymerase (E). In the presence of a fixed amount of sigma(70), the principal sigma for growth-related genes, the level of Esigma(70) holoenzyme formation increased linearly with the increase in core enzyme level, giving an apparent K:(d) for the core enzyme of 0.26 nM. Mixed reconstitution experiments in the presence of a fixed amount of core enzyme and increasing amounts of an equimolar mixture of all seven sigma subunits indicated that sigma(70) is strongest in terms of core enzyme binding, followed by sigma(N), sigma(F), sigma(E)/sigma(FecI), sigma(H) and sigma(S) in decreasing order. The orders of core binding affinity between sigma(70) and sigma(N) and between sigma(70) and sigma(H) were confirmed by measuring the replacement of one core-associated sigma by another sigma subunit. Taken together with the intracellular sigma levels, we tried to estimate the number of each holoenzyme form in growing E. coli cells.
Figures

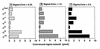

Similar articles
-
Two extracytoplasmic function sigma subunits, sigma(E) and sigma(FecI), of Escherichia coli: promoter selectivity and intracellular levels.J Bacteriol. 2000 Feb;182(4):1181-4. doi: 10.1128/JB.182.4.1181-1184.2000. J Bacteriol. 2000. PMID: 10648550 Free PMC article.
-
A mutation in region 1.1 of sigma70 affects promoter DNA binding by Escherichia coli RNA polymerase holoenzyme.EMBO J. 1999 Feb 1;18(3):709-16. doi: 10.1093/emboj/18.3.709. EMBO J. 1999. PMID: 9927430 Free PMC article.
-
The interaction between sigmaS, the stationary phase sigma factor, and the core enzyme of Escherichia coli RNA polymerase.Genes Cells. 2002 Mar;7(3):233-47. doi: 10.1046/j.1365-2443.2002.00517.x. Genes Cells. 2002. PMID: 11918668
-
How sigma docks to RNA polymerase and what sigma does.Curr Opin Microbiol. 2001 Apr;4(2):126-31. doi: 10.1016/s1369-5274(00)00177-6. Curr Opin Microbiol. 2001. PMID: 11282466 Review.
-
Functional modulation of Escherichia coli RNA polymerase.Annu Rev Microbiol. 2000;54:499-518. doi: 10.1146/annurev.micro.54.1.499. Annu Rev Microbiol. 2000. PMID: 11018136 Review.
Cited by
-
RNA polymerase approaches its promoter without long-range sliding along DNA.Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):9740-5. doi: 10.1073/pnas.1300221110. Epub 2013 May 29. Proc Natl Acad Sci U S A. 2013. PMID: 23720315 Free PMC article.
-
Coordinated Regulation of Rsd and RMF for Simultaneous Hibernation of Transcription Apparatus and Translation Machinery in Stationary-Phase Escherichia coli.Front Genet. 2019 Dec 4;10:1153. doi: 10.3389/fgene.2019.01153. eCollection 2019. Front Genet. 2019. PMID: 31867037 Free PMC article. Review.
-
Architecture of Burkholderia cepacia complex sigma70 gene family: evidence of alternative primary and clade-specific factors, and genomic instability.BMC Genomics. 2007 Sep 4;8:308. doi: 10.1186/1471-2164-8-308. BMC Genomics. 2007. PMID: 17784948 Free PMC article.
-
Trouble is coming: Signaling pathways that regulate general stress responses in bacteria.J Biol Chem. 2019 Aug 2;294(31):11685-11700. doi: 10.1074/jbc.REV119.005593. Epub 2019 Jun 13. J Biol Chem. 2019. PMID: 31197038 Free PMC article. Review.
-
Evidence for sigma factor competition in the regulation of alginate production by Pseudomonas aeruginosa.PLoS One. 2013 Aug 22;8(8):e72329. doi: 10.1371/journal.pone.0072329. eCollection 2013. PLoS One. 2013. PMID: 23991093 Free PMC article.
References
-
- Helmann J.D. and Chamberlin,M.J. (1988) Annu. Rev. Biochem., 57, 839–872. - PubMed
-
- Ishihama A. (1988) Trends Genet., 4, 282–286. - PubMed
-
- Gross C.A., Lonetto,M. and Losick,R. (1992) In Yamamoto,K. and McKnight,S. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 129–176.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

