Selective inhibition of splicing at the avian sarcoma virus src 3' splice site by direct-repeat posttranscriptional cis elements
- PMID: 10954552
- PMCID: PMC116363
- DOI: 10.1128/jvi.74.18.8513-8523.2000
Selective inhibition of splicing at the avian sarcoma virus src 3' splice site by direct-repeat posttranscriptional cis elements
Abstract
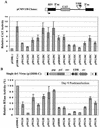
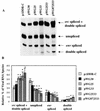
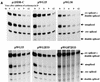
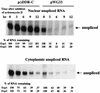
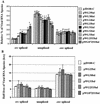
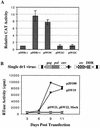
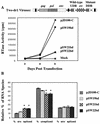
The direct-repeat elements (dr1) of avian sarcoma virus (ASV) and leukosis virus have the properties of constitutive transport elements (CTEs), which facilitate cytoplasmic accumulation of unspliced RNA. It is thought that these elements represent binding sites for cellular factors. Previous studies have indicated that in the context of the avian sarcoma virus genome, precise deletion of both ASV dr1 elements results in a very low level of virus replication. This is characterized by a decreased cytoplasmic accumulation of unspliced RNA and a selective increase in spliced src mRNA. Deletion of either the upstream or downstream dr1 results in a delayed-replication phenotype. To determine if the same regions of the dr1 mediate inhibition of src splicing and unspliced RNA transport, point mutations in the upstream and downstream elements were studied. In the context of viral genomes with single dr1 elements, the effects of the mutations on virus replication and increases in src splicing closely paralleled the effects of the mutations on CTE activity. For mutants strongly affecting CTE activity and splicing, unspliced RNA but not spliced RNA turned over in the nucleus more rapidly than wild-type RNA. In the context of wild-type virus containing two dr1 elements, mutations of either element that strongly affect CTE activity caused a marked delay of virus replication and a selective increase in src splicing. However, the turnover of the mutant unspliced RNA as well as the spliced mRNA species did not differ significantly from that of the wild type. These results suggest the dr1 elements in ASV act to selectively inhibit src splicing and that both elements contribute to the fitness of the wild-type virus. However, a single dr1 element is sufficient to stabilize unspliced RNA.
Figures


Similar articles
-
The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release.Virology. 1998 Jul 20;247(1):86-96. doi: 10.1006/viro.1998.9233. Virology. 1998. PMID: 9683574
-
Mutational analysis of the rous sarcoma virus DR posttranscriptional control element.J Virol. 1998 Apr;72(4):3407-11. doi: 10.1128/JVI.72.4.3407-3411.1998. J Virol. 1998. PMID: 9525671 Free PMC article.
-
Mutations in the regions of the Rous sarcoma virus 3' splice sites: implications for regulation of alternative splicing.J Virol. 1991 May;65(5):2640-6. doi: 10.1128/JVI.65.5.2640-2646.1991. J Virol. 1991. PMID: 1850037 Free PMC article.
-
Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication.Adv Virus Res. 2009;74:1-40. doi: 10.1016/S0065-3527(09)74001-1. Adv Virus Res. 2009. PMID: 19698894 Review.
-
Synthesis and processing of avian sarcoma retrovirus RNA.Adv Virus Res. 1988;35:1-38. doi: 10.1016/s0065-3527(08)60707-1. Adv Virus Res. 1988. PMID: 2852891 Review. No abstract available.
Cited by
-
Detection and molecular characterization of J subgroup avian leukosis virus in wild ducks in China.PLoS One. 2014 Apr 14;9(4):e94980. doi: 10.1371/journal.pone.0094980. eCollection 2014. PLoS One. 2014. PMID: 24733260 Free PMC article.
-
Repair of a Rev-minus human immunodeficiency virus type 1 mutant by activation of a cryptic splice site.J Virol. 2001 Apr;75(7):3495-500. doi: 10.1128/JVI.75.7.3495-3500.2001. J Virol. 2001. PMID: 11238879 Free PMC article.
-
A new RNA element located in the coding region of a murine endogenous retrovirus can functionally replace the Rev/Rev-responsive element system in human immunodeficiency virus type 1 Gag expression.J Virol. 2001 Nov;75(22):10670-82. doi: 10.1128/JVI.75.22.10670-10682.2001. J Virol. 2001. PMID: 11602709 Free PMC article.
-
RNA processing control in avian retroviruses.Front Biosci. 2008 May 1;13:3869-83. doi: 10.2741/2975. Front Biosci. 2008. PMID: 18508481 Free PMC article. Review.
-
Novel sequences of subgroup J avian leukosis viruses associated with hemangioma in Chinese layer hens.Virol J. 2011 Dec 21;8:552. doi: 10.1186/1743-422X-8-552. Virol J. 2011. PMID: 22185463 Free PMC article.
References
-
- Berberich S L, Stoltzfus C M. Analysis of spliced and unspliced Rous sarcoma virus RNAs early and late after infection of chicken embryo fibroblasts: effect of cell culture conditions. Virology. 1991;182:135–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

