In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632
- PMID: 10952574
- PMCID: PMC90064
- DOI: 10.1128/AAC.44.9.2319-2326.2000
In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632
Abstract
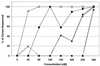
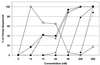
BMS-232632 is an azapeptide human immunodeficiency virus (HIV) type 1 (HIV-1) protease inhibitor that displays potent anti-HIV-1 activity (50% effective concentration [EC(50)], 2.6 to 5.3 nM; EC(90), 9 to 15 nM). In vitro passage of HIV-1 RF in the presence of inhibitors showed that BMS-232632 selected for resistant variants more slowly than nelfinavir or ritonavir did. Genotypic and phenotypic analysis of three different HIV strains resistant to BMS-232632 indicated that an N88S substitution in the viral protease appeared first during the selection process in two of the three strains. An I84V change appeared to be an important substitution in the third strain used. Mutations were also observed at the protease cleavage sites following drug selection. The evolution to resistance seemed distinct for each of the three strains used, suggesting multiple pathways to resistance and the importance of the viral genetic background. A cross-resistance study involving five other protease inhibitors indicated that BMS-232632-resistant virus remained sensitive to saquinavir, while it showed various levels (0. 1- to 71-fold decrease in sensitivity)-of cross-resistance to nelfinavir, indinavir, ritonavir, and amprenavir. In reciprocal experiments, the BMS-232632 susceptibility of HIV-1 variants selected in the presence of each of the other HIV-1 protease inhibitors showed that the nelfinavir-, saquinavir-, and amprenavir-resistant strains of HIV-1 remained sensitive to BMS-232632, while indinavir- and ritonavir-resistant viruses displayed six- to ninefold changes in BMS-232632 sensitivity. Taken together, our data suggest that BMS-232632 may be a valuable protease inhibitor for use in combination therapy.
Figures
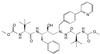

Similar articles
-
Activities of atazanavir (BMS-232632) against a large panel of human immunodeficiency virus type 1 clinical isolates resistant to one or more approved protease inhibitors.Antimicrob Agents Chemother. 2003 Apr;47(4):1324-33. doi: 10.1128/AAC.47.4.1324-1333.2003. Antimicrob Agents Chemother. 2003. PMID: 12654666 Free PMC article.
-
BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents.Antimicrob Agents Chemother. 2000 Aug;44(8):2093-9. doi: 10.1128/AAC.44.8.2093-2099.2000. Antimicrob Agents Chemother. 2000. PMID: 10898681 Free PMC article.
-
Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors.Proc Natl Acad Sci U S A. 1996 Feb 20;93(4):1648-53. doi: 10.1073/pnas.93.4.1648. Proc Natl Acad Sci U S A. 1996. PMID: 8643685 Free PMC article.
-
The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients.Antivir Ther. 2004 Jun;9(3):301-14. Antivir Ther. 2004. PMID: 15259893 Review.
-
Atazanavir/ritonavir: a review of its use in HIV therapy.Drugs Today (Barc). 2008 Feb;44(2):103-32. doi: 10.1358/dot.2008.44.2.1137107. Drugs Today (Barc). 2008. PMID: 18389089 Review.
Cited by
-
Atazanavir signature I50L resistance substitution accounts for unique phenotype of increased susceptibility to other protease inhibitors in a variety of human immunodeficiency virus type 1 genetic backbones.Antimicrob Agents Chemother. 2005 Sep;49(9):3816-24. doi: 10.1128/AAC.49.9.3816-3824.2005. Antimicrob Agents Chemother. 2005. PMID: 16127058 Free PMC article.
-
Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly.J Virol. 2002 Sep;76(17):8659-66. doi: 10.1128/jvi.76.17.8659-8666.2002. J Virol. 2002. PMID: 12163585 Free PMC article.
-
Mutations in multiple domains of Gag drive the emergence of in vitro resistance to the phosphonate-containing HIV-1 protease inhibitor GS-8374.J Virol. 2013 Jan;87(1):454-63. doi: 10.1128/JVI.01211-12. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097440 Free PMC article.
-
Atazanavir: in pediatric patients with HIV-1 infection.Paediatr Drugs. 2012 Apr 1;14(2):131-41. doi: 10.2165/11208550-000000000-00000. Paediatr Drugs. 2012. PMID: 22292486 Review.
-
Tenofovir comedication does not impair the steady-state pharmacokinetics of ritonavir-boosted atazanavir in HIV-1-infected adults.Eur J Clin Pharmacol. 2007 Oct;63(10):935-40. doi: 10.1007/s00228-007-0344-y. Epub 2007 Jul 31. Eur J Clin Pharmacol. 2007. PMID: 17665183 Clinical Trial.
References
-
- Cameron D W, Japour A J, Xu Y, Hsu A, Mellors J, Farthing C, Cohen C, Poretz D, Markowitz M, Follansbee S, Angel J B, McMahon D, Ho D, Devanarayan V, Rode R, Salgo M, Kempf D J, Granneman R, Leonard J M, Sun E. Ritonavir and saquinavir combination therapy for the treatment of HIV infection. AIDS. 1999;13:213–224. - PubMed
-
- Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–575. - PubMed
-
- Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

