Multiple splicing defects in an intronic false exon
- PMID: 10938119
- PMCID: PMC86117
- DOI: 10.1128/MCB.20.17.6414-6425.2000
Multiple splicing defects in an intronic false exon
Abstract
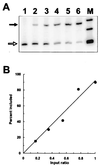
Splice site consensus sequences alone are insufficient to dictate the recognition of real constitutive splice sites within the typically large transcripts of higher eukaryotes, and large numbers of pseudoexons flanked by pseudosplice sites with good matches to the consensus sequences can be easily designated. In an attempt to identify elements that prevent pseudoexon splicing, we have systematically altered known splicing signals, as well as immediately adjacent flanking sequences, of an arbitrarily chosen pseudoexon from intron 1 of the human hprt gene. The substitution of a 5' splice site that perfectly matches the 5' consensus combined with mutation to match the CAG/G sequence of the 3' consensus failed to get this model pseudoexon included as the central exon in a dhfr minigene context. Provision of a real 3' splice site and a consensus 5' splice site and removal of an upstream inhibitory sequence were necessary and sufficient to confer splicing on the pseudoexon. This activated context also supported the splicing of a second pseudoexon sequence containing no apparent enhancer. Thus, both the 5' splice site sequence and the polypyrimidine tract of the pseudoexon are defective despite their good agreement with the consensus. On the other hand, the pseudoexon body did not exert a negative influence on splicing. The introduction into the pseudoexon of a sequence selected for binding to ASF/SF2 or its replacement with beta-globin exon 2 only partially reversed the effect of the upstream negative element and the defective polypyrimidine tract. These results support the idea that exon-bridging enhancers are not a prerequisite for constitutive exon definition and suggest that intrinsically defective splice sites and negative elements play important roles in distinguishing the real splicing signal from the vast number of false splicing signals.
Figures



Similar articles
-
Characterization of hprt splicing mutations induced by the ultimate carcinogenic metabolite of benzo[a]pyrene in Chinese hamster V-79 cells.Cancer Res. 1995 Apr 1;55(7):1550-8. Cancer Res. 1995. PMID: 7882364
-
Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells.Mol Cell Biol. 1993 Aug;13(8):5085-98. doi: 10.1128/mcb.13.8.5085-5098.1993. Mol Cell Biol. 1993. PMID: 8336736 Free PMC article.
-
An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing.J Biol Chem. 2004 Apr 23;279(17):16980-8. doi: 10.1074/jbc.M313439200. Epub 2004 Feb 13. J Biol Chem. 2004. PMID: 14966131
-
Pseudoexon activation in disease by non-splice site deep intronic sequence variation - wild type pseudoexons constitute high-risk sites in the human genome.Hum Mutat. 2022 Feb;43(2):103-127. doi: 10.1002/humu.24306. Epub 2021 Dec 5. Hum Mutat. 2022. PMID: 34837434 Review.
-
Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum.Mutat Res. 1998 Nov;411(3):179-214. doi: 10.1016/s1383-5742(98)00013-1. Mutat Res. 1998. PMID: 9804951 Review.
Cited by
-
Dual role of G-runs and hnRNP F in the regulation of a mutation-activated pseudoexon in the fibrinogen gamma-chain transcript.PLoS One. 2013;8(3):e59333. doi: 10.1371/journal.pone.0059333. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533617 Free PMC article.
-
Antisense Oligonucleotides Modulating Activation of a Nonsense-Mediated RNA Decay Switch Exon in the ATM Gene.Nucleic Acid Ther. 2016 Dec;26(6):392-400. doi: 10.1089/nat.2016.0635. Epub 2016 Sep 22. Nucleic Acid Ther. 2016. PMID: 27658045 Free PMC article.
-
Global identification of hnRNP A1 binding sites for SSO-based splicing modulation.BMC Biol. 2016 Jul 5;14:54. doi: 10.1186/s12915-016-0279-9. BMC Biol. 2016. PMID: 27380775 Free PMC article.
-
A day in the life of the spliceosome.Nat Rev Mol Cell Biol. 2014 Feb;15(2):108-21. doi: 10.1038/nrm3742. Nat Rev Mol Cell Biol. 2014. PMID: 24452469 Free PMC article. Review.
-
Novel Splicing of Immune System Thyroid Stimulating Hormone β-Subunit-Genetic Regulation and Biological Importance.Front Endocrinol (Lausanne). 2019 Feb 5;10:44. doi: 10.3389/fendo.2019.00044. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 30804891 Free PMC article. Review.
References
-
- Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. - PubMed
-
- Balvay L, Libri D, Fiszman M Y. Pre-mRNA secondary structure and the regulation of splicing. Bioessays. 1993;15:165–169. - PubMed
-
- Batzer M A, Deininger P L, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin C M, Schmid C W, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. - PubMed
-
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
