The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5' splice site
- PMID: 10938105
- PMCID: PMC86103
- DOI: 10.1128/MCB.20.17.6287-6299.2000
The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5' splice site
Abstract
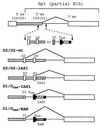
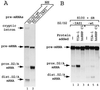
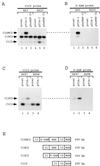
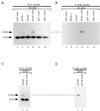
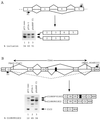
Splicing of the K-SAM alternative exon of the fibroblast growth factor receptor 2 gene is heavily dependent on the U-rich sequence IAS1 lying immediately downstream from its 5' splice site. We show that IAS1 can activate the use of several heterologous 5' splice sites in vitro. Addition of the RNA-binding protein TIA-1 to splicing extracts preferentially enhances the use of 5' splice sites linked to IAS1. TIA-1 can provoke a switch to use of such sites on pre-mRNAs with competing 5' splice sites, only one of which is adjacent to IAS1. Using a combination of UV cross-linking and specific immunoprecipitation steps, we show that TIA-1 binds to IAS1 in cell extracts. This binding is stronger if IAS1 is adjacent to a 5' splice site and is U1 snRNP dependent. Overexpression of TIA-1 in cultured cells activates K-SAM exon splicing in an IAS1-dependent manner. If IAS1 is replaced with a bacteriophage MS2 operator, splicing of the K-SAM exon can no longer be activated by TIA-1. Splicing can, however, be activated by a TIA-1-MS2 coat protein fusion, provided that the operator is close to the 5' splice site. Our results identify TIA-1 as a novel splicing regulator, which acts by binding to intron sequences immediately downstream from a 5' splice site in a U1 snRNP-dependent fashion. TIA-1 is distantly related to the yeast U1 snRNP protein Nam8p, and the functional similarities between the two proteins are discussed.
Figures


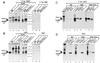

Similar articles
-
Cooperative binding of TIA-1 and U1 snRNP in K-SAM exon splicing activation.Biochem Biophys Res Commun. 2007 Jul 13;358(4):1065-70. doi: 10.1016/j.bbrc.2007.05.050. Epub 2007 May 15. Biochem Biophys Res Commun. 2007. PMID: 17512901
-
An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing.J Biol Chem. 2004 Apr 23;279(17):16980-8. doi: 10.1074/jbc.M313439200. Epub 2004 Feb 13. J Biol Chem. 2004. PMID: 14966131
-
The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing.Mol Cell. 2000 Nov;6(5):1089-98. doi: 10.1016/s1097-2765(00)00107-6. Mol Cell. 2000. PMID: 11106748
-
Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1.Apoptosis. 2001 Dec;6(6):463-8. doi: 10.1023/a:1012441824719. Apoptosis. 2001. PMID: 11595836 Review.
-
Mechanisms for selecting 5' splice sites in mammalian pre-mRNA splicing.Trends Genet. 1994 Mar;10(3):100-6. doi: 10.1016/0168-9525(94)90233-x. Trends Genet. 1994. PMID: 8178363 Review.
Cited by
-
T-cell intracellular antigens function as tumor suppressor genes.Cell Death Dis. 2015 Mar 5;6(3):e1669. doi: 10.1038/cddis.2015.43. Cell Death Dis. 2015. PMID: 25741594 Free PMC article.
-
Frequent gain and loss of intronic splicing regulatory elements during the evolution of vertebrates.Genome Biol Evol. 2012;4(7):659-74. doi: 10.1093/gbe/evs051. Epub 2012 May 21. Genome Biol Evol. 2012. PMID: 22619362 Free PMC article.
-
Two predicted α-helices within the prion-like domain of TIAR-1 play a crucial role in its association with stress granules in Caenorhabditis elegans.Front Cell Dev Biol. 2023 Dec 15;11:1265104. doi: 10.3389/fcell.2023.1265104. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38161334 Free PMC article.
-
Binding of hnRNP H and U2AF65 to respective G-codes and a poly-uridine tract collaborate in the N50-5'ss selection of the REST N exon in H69 cells.PLoS One. 2012;7(7):e40315. doi: 10.1371/journal.pone.0040315. Epub 2012 Jul 5. PLoS One. 2012. PMID: 22792276 Free PMC article.
-
Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules.Mol Biol Cell. 2002 Jan;13(1):195-210. doi: 10.1091/mbc.01-05-0221. Mol Biol Cell. 2002. PMID: 11809833 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
