Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells
- PMID: 10933723
- PMCID: PMC112346
- DOI: 10.1128/jvi.74.17.8127-8134.2000
Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells
Abstract
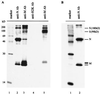
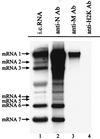
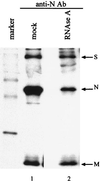
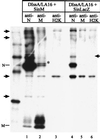
Coronavirus contains three envelope proteins, M, E and S, and a nucleocapsid, which consists of genomic RNA and N protein, within the viral envelope. We studied the macromolecular interactions involved in coronavirus assembly in cells infected with a murine coronavirus, mouse hepatitis virus (MHV). Coimmunoprecipitation analyses demonstrated an interaction between N protein and M protein in infected cells. Pulse-labeling experiments showed that newly synthesized, unglycosylated M protein interacted with N protein in a pre-Golgi compartment, which is part of the MHV budding site. Coimmunoprecipitation analyses further revealed that M protein interacted with only genomic-length MHV mRNA, mRNA 1, while N protein interacted with all MHV mRNAs. These data indicated that M protein interacted with the nucleocapsid, consisting of N protein and mRNA 1, in infected cells. The M protein-nucleocapsid interaction occurred in the absence of S and E proteins. Intracellular M protein-N protein interaction was maintained after removal of viral RNAs by RNase treatment. However, the M protein-N protein interaction did not occur in cells coexpressing M protein and N protein alone. These data indicated that while the M protein-N protein interaction, which is independent of viral RNA, occurred in the M protein-nucleocapsid complex, some MHV function(s) was necessary for the initiation of M protein-nucleocapsid interaction. The M protein-nucleocapsid interaction, which occurred near or at the MHV budding site, most probably represented the process of specific packaging of the MHV genome into MHV particles.
Figures


Similar articles
-
Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus.J Virol. 2002 May;76(10):4987-99. doi: 10.1128/jvi.76.10.4987-4999.2002. J Virol. 2002. PMID: 11967315 Free PMC article.
-
Characterization of N protein self-association in coronavirus ribonucleoprotein complexes.Virus Res. 2003 Dec;98(2):131-40. doi: 10.1016/j.virusres.2003.08.021. Virus Res. 2003. PMID: 14659560 Free PMC article.
-
Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras.J Virol. 2016 Apr 14;90(9):4357-4368. doi: 10.1128/JVI.03212-15. Print 2016 May. J Virol. 2016. PMID: 26889024 Free PMC article.
-
[Coronaviruses].Uirusu. 2011 Dec;61(2):205-10. doi: 10.2222/jsv.61.205. Uirusu. 2011. PMID: 22916567 Review. Japanese.
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
Cited by
-
The SARS-CoV-2 nucleoprotein associates with anionic lipid membranes.J Biol Chem. 2024 Aug;300(8):107456. doi: 10.1016/j.jbc.2024.107456. Epub 2024 Jun 10. J Biol Chem. 2024. PMID: 38866325 Free PMC article.
-
COVID-19 Variants and Vaccine Development.Viruses. 2024 May 10;16(5):757. doi: 10.3390/v16050757. Viruses. 2024. PMID: 38793638 Free PMC article. Review.
-
Detection of SARS-CoV-2 N protein using AgNPs-modified aligned silicon nanowires BioSERS chip.RSC Adv. 2024 Apr 16;14(17):12071-12080. doi: 10.1039/d4ra00267a. eCollection 2024 Apr 10. RSC Adv. 2024. PMID: 38628480 Free PMC article.
-
SARS-CoV-2 outbreak: role of viral proteins and genomic diversity in virus infection and COVID-19 progression.Virol J. 2024 Mar 27;21(1):75. doi: 10.1186/s12985-024-02342-w. Virol J. 2024. PMID: 38539202 Free PMC article. Review.
-
A Comprehensive View on the Protein Functions of Porcine Epidemic Diarrhea Virus.Genes (Basel). 2024 Jan 26;15(2):165. doi: 10.3390/genes15020165. Genes (Basel). 2024. PMID: 38397155 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

