Speeding molecular recognition by using the folding funnel: the fly-casting mechanism
- PMID: 10908673
- PMCID: PMC16787
- DOI: 10.1073/pnas.160259697
Speeding molecular recognition by using the folding funnel: the fly-casting mechanism
Abstract
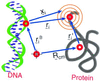
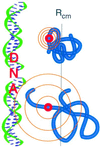
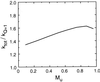
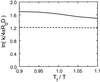
Protein folding and binding are kindred processes. Many proteins in the cell are unfolded, so folding and function are coupled. This paper investigates how binding kinetics is influenced by the folding of a protein. We find that a relatively unstructured protein molecule can have a greater capture radius for a specific binding site than the folded state with its restricted conformational freedom. In this scenario of binding, the unfolded state binds weakly at a relatively large distance followed by folding as the protein approaches the binding site: the "fly-casting mechanism." We illustrate this scenario with the hypothetical kinetics of binding a single repressor molecule to a DNA site and find that the binding rate can be significantly enhanced over the rate of binding of a fully folded protein.
Figures


Similar articles
-
Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the "fly-casting" mechanism.J Mol Biol. 2009 Nov 13;393(5):1143-59. doi: 10.1016/j.jmb.2009.09.010. Epub 2009 Sep 10. J Mol Biol. 2009. PMID: 19747922
-
Folding of Desulfovibrio desulfuricans flavodoxin is accelerated by cofactor fly-casting.Arch Biochem Biophys. 2006 Jul 1;451(1):51-8. doi: 10.1016/j.abb.2006.03.032. Epub 2006 Apr 21. Arch Biochem Biophys. 2006. PMID: 16730634
-
Mechanism of coupled folding and binding of an intrinsically disordered protein.Nature. 2007 Jun 21;447(7147):1021-5. doi: 10.1038/nature05858. Epub 2007 May 23. Nature. 2007. PMID: 17522630
-
Coupled folding and specific binding: fishing for amphiphilicity.Int J Mol Sci. 2011;12(3):1431-50. doi: 10.3390/ijms12031431. Epub 2011 Feb 24. Int J Mol Sci. 2011. PMID: 21673899 Free PMC article. Review.
-
Dynamics and mechanisms of coupled protein folding and binding reactions.Curr Opin Struct Biol. 2012 Feb;22(1):21-9. doi: 10.1016/j.sbi.2011.09.010. Epub 2011 Nov 29. Curr Opin Struct Biol. 2012. PMID: 22129832 Review.
Cited by
-
A comparative study of protein-ssDNA interactions.NAR Genom Bioinform. 2021 Feb 23;3(1):lqab006. doi: 10.1093/nargab/lqab006. eCollection 2021 Mar. NAR Genom Bioinform. 2021. PMID: 33655206 Free PMC article.
-
Binding Mechanisms of Intrinsically Disordered Proteins: Theory, Simulation, and Experiment.Front Mol Biosci. 2016 Sep 9;3:52. doi: 10.3389/fmolb.2016.00052. eCollection 2016. Front Mol Biosci. 2016. PMID: 27668217 Free PMC article. Review.
-
Substrate-specific reorganization of the conformational ensemble of CSK implicates novel modes of kinase function.PLoS Comput Biol. 2012;8(9):e1002695. doi: 10.1371/journal.pcbi.1002695. Epub 2012 Sep 20. PLoS Comput Biol. 2012. PMID: 23028292 Free PMC article.
-
Inadequate Reference Datasets Biased toward Short Non-epitopes Confound B-cell Epitope Prediction.J Biol Chem. 2016 Jul 8;291(28):14585-99. doi: 10.1074/jbc.M116.729020. Epub 2016 May 9. J Biol Chem. 2016. PMID: 27189949 Free PMC article.
-
Mutual synergistic protein folding in split intein.Biosci Rep. 2012 Oct;32(5):433-42. doi: 10.1042/BSR20120049. Biosci Rep. 2012. PMID: 22681309 Free PMC article.
References
-
- Wright P E, Dyson H J. J Mol Biol. 1999;293:321–331. - PubMed
-
- Ptashne M. A Genetic Switch: Phage λ and Higher Organisms. 2nd Ed. Cambridge, MA: Blackwell Scientific; 1992.
-
- Frauenfelder H, Sligar S G, Wolynes P G. Science. 1991;254:1598–1603. - PubMed
-
- Frauenfelder H, Petsko G A, Tsernoglou D. Nature (London) 1979;280:558–563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

