Patterns of variant polyadenylation signal usage in human genes
- PMID: 10899149
- PMCID: PMC310884
- DOI: 10.1101/gr.10.7.1001
Patterns of variant polyadenylation signal usage in human genes
Abstract
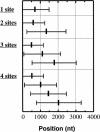
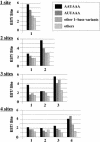
The formation of mature mRNAs in vertebrates involves the cleavage and polyadenylation of the pre-mRNA, 10-30 nt downstream of an AAUAAA or AUUAAA signal sequence. The extensive cDNA data now available shows that these hexamers are not strictly conserved. In order to identify variant polyadenylation signals on a large scale, we compared over 8700 human 3' untranslated sequences to 157,775 polyadenylated expressed sequence tags (ESTs), used as markers of actual mRNA 3' ends. About 5600 EST-supported putative mRNA 3' ends were collected and analyzed for significant hexameric sequences. Known polyadenylation signals were found in only 73% of the 3' fragments. Ten single-base variants of the AAUAAA sequence were identified with a highly significant occurrence rate, potentially representing 14.9% of the actual polyadenylation signals. Of the mRNAs, 28.6% displayed two or more polyadenylation sites. In these mRNAs, the poly(A) sites proximal to the coding sequence tend to use variant signals more often, while the 3'-most site tends to use a canonical signal. The average number of ESTs associated with each signal type suggests that variant signals (including the common AUUAAA) are processed less efficiently than the canonical signal and could therefore be selected for regulatory purposes. However, the position of the site in the untranslated region may also play a role in polyadenylation rate.
Figures


Similar articles
-
Identification and characterization of polyadenylation signal (PAS) variants in human genomic sequences based on modified EST clustering.In Silico Biol. 2008;8(3-4):347-61. In Silico Biol. 2008. PMID: 19032167
-
Similarities and differences of polyadenylation signals in human and fly.BMC Genomics. 2006 Jul 12;7:176. doi: 10.1186/1471-2164-7-176. BMC Genomics. 2006. PMID: 16836751 Free PMC article.
-
Systematic variation in mRNA 3'-processing signals during mouse spermatogenesis.Nucleic Acids Res. 2007;35(1):234-46. doi: 10.1093/nar/gkl919. Epub 2006 Dec 8. Nucleic Acids Res. 2007. PMID: 17158511 Free PMC article.
-
3'-End processing of pre-mRNA in eukaryotes.FEMS Microbiol Rev. 1999 Jun;23(3):277-95. doi: 10.1111/j.1574-6976.1999.tb00400.x. FEMS Microbiol Rev. 1999. PMID: 10371034 Review.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
-
Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta.BMC Evol Biol. 2015 Jul 30;15:148. doi: 10.1186/s12862-015-0431-0. BMC Evol Biol. 2015. PMID: 26223266 Free PMC article.
-
Enrichment Reveals Extensive Integration of Hepatitis B Virus DNA in Hepatitis Delta Virus-Infected Patients.J Infect Dis. 2024 Sep 23;230(3):e684-e693. doi: 10.1093/infdis/jiae045. J Infect Dis. 2024. PMID: 38271697 Free PMC article.
-
Re-editing the paradigm of Cytidine (C) to Uridine (U) RNA editing.RNA Biol. 2014;11(10):1233-7. doi: 10.1080/15476286.2014.996054. RNA Biol. 2014. PMID: 25585043 Free PMC article.
-
Two newly identified exons in human GRM1 express a novel splice variant of metabotropic glutamate 1 receptor.Gene. 2013 May 1;519(2):367-73. doi: 10.1016/j.gene.2013.02.009. Epub 2013 Feb 24. Gene. 2013. PMID: 23481697 Free PMC article.
-
Analysis of Polyadenylation Signal Usage with Full-Length Transcriptome in Spodoptera frugiperda (Lepidoptera: Noctuidae).Insects. 2022 Sep 2;13(9):803. doi: 10.3390/insects13090803. Insects. 2022. PMID: 36135504 Free PMC article.
References
-
- Anand S, Batista FD, Tkach T, Efremov DG, Burrone OR. Multiple transcripts of the murine immunoglobulin epsilon membrane locus are generated by alternative splicing and differential usage of two polyadenylation sites. Mol Immunol. 1997;34:175–183. - PubMed
-
- Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM, Harmar AJ. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72:1384–1388. - PubMed
-
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for expressed sequence tags. Nat Genet. 1993;4:332–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
