Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya
- PMID: 10878065
- PMCID: PMC87000
- DOI: 10.1128/JCM.38.7.2688-2695.2000
Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya
Abstract
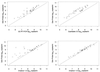
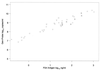
Accurate and sensitive quantification of human immunodeficiency virus type 1 (HIV-1) RNA has been invaluable as a marker for disease prognosis and for clinical monitoring of HIV-1 disease. The first generation of commercially available HIV-1 RNA tests were optimized to detect the predominant HIV-1 subtype found in North America and Europe, subtype B. However, these tests are frequently suboptimal in detecting HIV-1 genetic forms or subtypes found in other parts of the world. The goal of the present study was to evaluate the performance of a new viral load assay with non-subtype B viruses. A transcription-mediated amplification method for detection and quantitation of diverse HIV-1 subtypes, called the Gen-Probe HIV-1 viral load assay, is under development. In this study we examined the performance of the Gen-Probe HIV-1 viral load assay relative to that of the commonly used commercial HIV-1 RNA assays using a panel of primary isolates from Kenya. For comparison, we included several subtype B cloned viruses, and we quantified each virus using an in-house quantitative-competitive reverse transcriptase PCR (QC-RT-PCR) method and gag(p24) antigen capture. The Gen-Probe HIV-1 viral load assay and a version of the Roche AMPLICOR HIV-1 MONITOR test (version 1.5) that was designed to detect a broader range of subtypes were both sensitive for the quantification of Kenyan primary isolates, which represented subtype A, C, and D viruses. The Gen-Probe HIV-1 viral load assay was more sensitive for the majority of viruses than the Roche AMPLICOR HIV-1 MONITOR test version 1.0, the Bayer Quantiplex HIV RNA 3.0 assay, or a QC-RT-PCR method in use in our laboratory, suggesting that it provides a useful method for quantifying HIV-1 RNAs from diverse parts of the world, including Africa.
Figures

Similar articles
-
Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0.J Clin Microbiol. 2001 Mar;39(3):862-70. doi: 10.1128/JCM.39.3.862-870.2001. J Clin Microbiol. 2001. PMID: 11230396 Free PMC article.
-
Evaluation of two commercially available, inexpensive alternative assays used for assessing viral load in a cohort of human immunodeficiency virus type 1 subtype C-infected patients from South Africa.J Clin Microbiol. 2005 Feb;43(2):857-61. doi: 10.1128/JCM.43.2.857-861.2005. J Clin Microbiol. 2005. PMID: 15695692 Free PMC article.
-
Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknika NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma.J Clin Microbiol. 2000 Nov;38(11):4034-41. doi: 10.1128/JCM.38.11.4034-4041.2000. J Clin Microbiol. 2000. PMID: 11060065 Free PMC article.
-
The measurement of HIV-1 viral load in resource-limited settings: how and where?Clin Lab. 2007;53(3-4):135-48. Clin Lab. 2007. PMID: 17447649 Review.
-
Measurement of HIV-1 p24 antigen by signal-amplification-boosted ELISA of heat-denatured plasma is a simple and inexpensive alternative to tests for viral RNA.AIDS Rev. 2002 Apr-Jun;4(2):83-92. AIDS Rev. 2002. PMID: 12152521 Review.
Cited by
-
Breast milk cellular HIV-specific interferon γ responses are associated with protection from peripartum HIV transmission.AIDS. 2012 Oct 23;26(16):2007-16. doi: 10.1097/QAD.0b013e328359b7e0. AIDS. 2012. PMID: 22948269 Free PMC article.
-
HIV-exposed uninfected infants: elevated cord blood Interleukin 8 (IL-8) is significantly associated with maternal HIV infection and systemic IL-8 in a Kenyan cohort.Clin Transl Med. 2018 Sep 10;7(1):26. doi: 10.1186/s40169-018-0206-5. Clin Transl Med. 2018. PMID: 30198049 Free PMC article.
-
Depot Medroxyprogesterone Acetate Use Is Associated With Elevated Innate Immune Effector Molecules in Cervicovaginal Secretions of HIV-1-Uninfected Women.J Acquir Immune Defic Syndr. 2015 May 1;69(1):1-10. doi: 10.1097/QAI.0000000000000533. J Acquir Immune Defic Syndr. 2015. PMID: 25622059 Free PMC article.
-
Hormonal contraception and HIV-1 disease progression among postpartum Kenyan women.AIDS. 2007 Mar 30;21(6):749-53. doi: 10.1097/QAD.0b013e328032790f. AIDS. 2007. PMID: 17413696 Free PMC article.
-
Validation of performance of the gen-probe human immunodeficiency virus type 1 viral load assay with genital swabs and breast milk samples.J Clin Microbiol. 2002 Nov;40(11):3929-37. doi: 10.1128/JCM.40.11.3929-3937.2002. J Clin Microbiol. 2002. PMID: 12409354 Free PMC article.
References
-
- Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. - PubMed
-
- Berger A, Braner J, Doerr H W, Weber B. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus and hepatitis C virus infection. Intervirology. 1998;41:24–34. - PubMed
-
- Bodrug S, Domingo R, Holloway J, Sanders M, Nunomura K, Sloan C, Billyard B. Gen-Probe single tube quantitative HIV assay. J Clin Microbiol Infect. 1997;3:1050.
-
- Brodine S K, Mascola J R, Weiss P J, Ito S I, Porter K R, Artenstein A W, Garland F C, McCutchan F E, Burke D S. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet. 1995;346:1198–1199. - PubMed
-
- Brodine S K, Shaffer R A, Starkey M J, Tasker S A, Gilcrest J L, Louder M K, Barile A, VanCott T C, Vahey M T, McCutchan F E, Birx D L, Richman D D, Mascola J R. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann Intern Med. 1999;131:502–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

