Analysis of uracil-DNA glycosylases from the murine Ung gene reveals differential expression in tissues and in embryonic development and a subcellular sorting pattern that differs from the human homologues
- PMID: 10871356
- PMCID: PMC102736
- DOI: 10.1093/nar/28.12.2277
Analysis of uracil-DNA glycosylases from the murine Ung gene reveals differential expression in tissues and in embryonic development and a subcellular sorting pattern that differs from the human homologues
Abstract
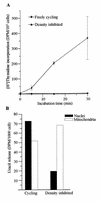
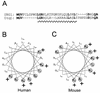
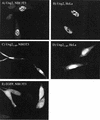
The murine UNG: gene encodes both mitochondrial (Ung1) and nuclear (Ung2) forms of uracil-DNA glyco-sylase. The gene contains seven exons organised like the human counterpart. While the putative Ung1 promoter (P(B)) and the human P(B) contain essentially the same, although differently organised, transcription factor binding elements, the Ung2 promoter (P(A)) shows limited homology to the human counterpart. Transient transfection of chimaeric promoter-luciferase constructs demonstrated that both promoters are functional and that P(B) drives transcription more efficiently than P(A). mRNAs for Ung1 and Ung2 are found in all adult tissues analysed, but they are differentially expressed. Furthermore, transcription of both mRNA forms, particularly Ung2, is induced in mid-gestation embryos. Except for a strong conservation of the 26 N-terminal residues in Ung2, the subcellular targeting sequences in the encoded proteins have limited homology. Ung2 is transported exclusively to the nucleus in NIH 3T3 cells as expected. In contrast, Ung1 was sorted both to nuclei and mitochondria. These results demonstrate that although the catalytic domain of uracil-DNA glycosylase is highly conserved in mouse and man, regulatory elements in the gene and subcellular sorting sequences in the proteins differ both structurally and functionally, resulting in altered contribution of the isoforms to total uracil-DNA glycosylase activity.
Figures




Similar articles
-
Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively.Nucleic Acids Res. 1998 Oct 15;26(20):4611-7. doi: 10.1093/nar/26.20.4611. Nucleic Acids Res. 1998. PMID: 9753728 Free PMC article.
-
Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene.Nucleic Acids Res. 1997 Feb 15;25(4):750-5. doi: 10.1093/nar/25.4.750. Nucleic Acids Res. 1997. PMID: 9016624 Free PMC article.
-
Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase.Nucleic Acids Res. 1998 Mar 15;26(6):1449-57. doi: 10.1093/nar/26.6.1449. Nucleic Acids Res. 1998. PMID: 9490791 Free PMC article.
-
Properties and functions of human uracil-DNA glycosylase from the UNG gene.Prog Nucleic Acid Res Mol Biol. 2001;68:365-86. doi: 10.1016/s0079-6603(01)68112-1. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554311 Review.
-
The nature of enzymes involved in uracil-DNA repair: isoform characteristics of proteins responsible for nuclear and mitochondrial genomic integrity.Curr Protein Pept Sci. 2001 Dec;2(4):335-47. doi: 10.2174/1389203013381044. Curr Protein Pept Sci. 2001. PMID: 12369930 Review.
Cited by
-
Uracil-DNA glycosylase UNG1 isoform variant supports class switch recombination and repairs nuclear genomic uracil.Nucleic Acids Res. 2019 May 21;47(9):4569-4585. doi: 10.1093/nar/gkz145. Nucleic Acids Res. 2019. PMID: 30838409 Free PMC article.
-
Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA.J Biol Chem. 2008 Sep 5;283(36):25046-56. doi: 10.1074/jbc.M803776200. Epub 2008 Jul 3. J Biol Chem. 2008. PMID: 18603530 Free PMC article.
-
UNG protects B cells from AID-induced telomere loss.J Exp Med. 2016 Oct 17;213(11):2459-2472. doi: 10.1084/jem.20160635. Epub 2016 Oct 3. J Exp Med. 2016. PMID: 27697833 Free PMC article.
-
Mitochondrial DNA toxicity in forebrain neurons causes apoptosis, neurodegeneration, and impaired behavior.Mol Cell Biol. 2010 Mar;30(6):1357-67. doi: 10.1128/MCB.01149-09. Epub 2010 Jan 11. Mol Cell Biol. 2010. PMID: 20065039 Free PMC article.
-
Highly efficient base excision repair (BER) in human and rat male germ cells.Nucleic Acids Res. 2001 Apr 15;29(8):1781-90. doi: 10.1093/nar/29.8.1781. Nucleic Acids Res. 2001. PMID: 11292851 Free PMC article.
References
-
- Lindahl T. (1993) Nature, 362, 709–715. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

