Orphan receptor DAX-1 is a shuttling RNA binding protein associated with polyribosomes via mRNA
- PMID: 10848616
- PMCID: PMC85942
- DOI: 10.1128/MCB.20.13.4910-4921.2000
Orphan receptor DAX-1 is a shuttling RNA binding protein associated with polyribosomes via mRNA
Abstract
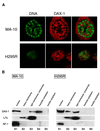
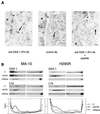
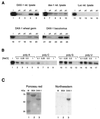
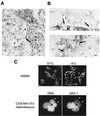
The DAX-1 (NR0B1) gene encodes an unusual member of the nuclear hormone receptor superfamily which acts as a transcriptional repressor. Mutations in the human DAX-1 gene cause X-linked adrenal hypoplasia congenita (AHC) associated with hypogonadotropic hypogonadism (HHG). We have studied the intracellular localization of the DAX-1 protein in human adrenal cortex and mouse Leydig tumor cells and found it to be both nuclear and cytoplasmic. A significant proportion of DAX-1 is associated with polyribosomes and is found complexed with polyadenylated RNA. DAX-1 directly binds to RNA, two domains within the protein being responsible for cooperative binding activity and specificity. Mutations in DAX-1 found in AHC-HHG patients significantly impair RNA binding. These findings reveal that DAX-1 plays multiple regulatory roles at the transcriptional and posttranscriptional levels.
Figures



Similar articles
-
X-linked adrenal hypoplasia congenita is caused by abnormal nuclear localization of the DAX-1 protein.Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):8225-30. doi: 10.1073/pnas.122044099. Epub 2002 May 28. Proc Natl Acad Sci U S A. 2002. PMID: 12034880 Free PMC article.
-
Structure-function analysis reveals the molecular determinants of the impaired biological function of DAX-1 mutants in AHC patients.Hum Mol Genet. 2003 May 1;12(9):1063-72. doi: 10.1093/hmg/ddg108. Hum Mol Genet. 2003. PMID: 12700175
-
DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita.Mol Cell Biol. 1997 Mar;17(3):1476-83. doi: 10.1128/MCB.17.3.1476. Mol Cell Biol. 1997. PMID: 9032275 Free PMC article.
-
The gene responsible for adrenal hypoplasia congenita, DAX-1, encodes a nuclear hormone receptor that defines a new class within the superfamily.Recent Prog Horm Res. 1996;51:241-59; discussion 259-60. Recent Prog Horm Res. 1996. PMID: 8701082 Review.
-
[DAX-1 abnormality].Nihon Rinsho. 2002 Feb;60(2):391-6. Nihon Rinsho. 2002. PMID: 11857932 Review. Japanese.
Cited by
-
Developmental expression of DAX1 in the European sea bass, Dicentrarchus labrax: lack of evidence for sexual dimorphism during sex differentiation.Reprod Biol Endocrinol. 2007 May 30;5:19. doi: 10.1186/1477-7827-5-19. Reprod Biol Endocrinol. 2007. PMID: 17537257 Free PMC article.
-
Orphan nuclear receptor DAX-1 in human endometrium and its disorders.Cancer Sci. 2005 Oct;96(10):645-52. doi: 10.1111/j.1349-7006.2005.00101.x. Cancer Sci. 2005. PMID: 16232195 Free PMC article.
-
Heat shock affects trafficking of DAX-1 by inducing its rapid and reversible cytoplasmic localization.Endocrine. 2005 Nov;28(2):137-44. doi: 10.1385/ENDO:28:2:137. Endocrine. 2005. PMID: 16388085
-
Role of dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1 in protein kinase A- and protein kinase C-mediated regulation of the steroidogenic acute regulatory protein expression in mouse Leydig tumor cells: mechanism of action.Endocrinology. 2009 Jan;150(1):187-99. doi: 10.1210/en.2008-0368. Epub 2008 Sep 11. Endocrinology. 2009. PMID: 18787026 Free PMC article.
-
X-linked adrenal hypoplasia congenita is caused by abnormal nuclear localization of the DAX-1 protein.Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):8225-30. doi: 10.1073/pnas.122044099. Epub 2002 May 28. Proc Natl Acad Sci U S A. 2002. PMID: 12034880 Free PMC article.
References
-
- Bae D S, Schaefer M L, Partan B W, Muglia L. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology. 1996;137:3921–3927. - PubMed
-
- Bardoni B, Zanaria E, Guioli S, Floridia G, Worley K C, Tonini G, Ferrante E, Chiumello G, McCabe E R B, Fraccaro M, Zuffardi O, Camerino G. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. - PubMed
-
- Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. - PubMed
-
- Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum Mol Genet. 1999;8:775–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
