Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes
- PMID: 10835360
- PMCID: PMC212761
- DOI: 10.1093/emboj/19.11.2629
Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes
Abstract
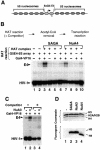
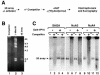
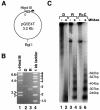
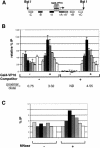
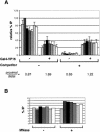
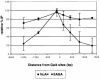
We analyzed the targeting of histone acetyltransferase (HAT) complexes by DNA-binding activators during transcriptional activation and the resulting distribution of acetylated histones. An in vitro competition assay was developed to acetylate and transcribe a nucleosomal array template in the presence of excess non-specific chromatin, which mimics in vivo conditions. Stimulation of transcription from the nucleosomal array template under competitive conditions by the SAGA and NuA4 HAT complexes depended on the presence of the Gal4-VP16 activator, which recognizes sites in the promoter and directly interacts with these HATs. Importantly, the stimulation of transcription by SAGA and NuA4 depended on the presence of Gal4-VP16 during histone acetylation, and Gal4-VP16-bound nucleosomal templates were acetylated preferentially by SAGA and NuA4 relative to the competitor chromatin. While targeting of the SAGA complex led to H3 acetylation of promoter-proximal nucleosomes, targeting of the NuA4 complex led to a broader domain of H4 acetylation of >3 kbp. Thus, either promoter-proximal H3 acetylation by SAGA or broadly distributed acetylation of H4 by NuA4 activated transcription from chromatin templates.
Figures



Similar articles
-
Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation.EMBO J. 2004 Jul 7;23(13):2597-607. doi: 10.1038/sj.emboj.7600230. Epub 2004 Jun 3. EMBO J. 2004. PMID: 15175650 Free PMC article.
-
NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p.EMBO J. 1999 Sep 15;18(18):5108-19. doi: 10.1093/emboj/18.18.5108. EMBO J. 1999. PMID: 10487762 Free PMC article.
-
Transcriptional activators direct histone acetyltransferase complexes to nucleosomes.Nature. 1998 Jul 30;394(6692):498-502. doi: 10.1038/28886. Nature. 1998. PMID: 9697775
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
Histone acetyltransferase complexes.Semin Cell Dev Biol. 1999 Apr;10(2):169-77. doi: 10.1006/scdb.1999.0298. Semin Cell Dev Biol. 1999. PMID: 10441070 Review.
Cited by
-
Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing.EMBO J. 2001 Oct 15;20(20):5759-68. doi: 10.1093/emboj/20.20.5759. EMBO J. 2001. PMID: 11598018 Free PMC article.
-
Global position and recruitment of HATs and HDACs in the yeast genome.Mol Cell. 2004 Oct 22;16(2):199-209. doi: 10.1016/j.molcel.2004.09.021. Mol Cell. 2004. PMID: 15494307 Free PMC article.
-
Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration.Proc Natl Acad Sci U S A. 2005 Jun 14;102(24):8472-7. doi: 10.1073/pnas.0503505102. Epub 2005 Jun 2. Proc Natl Acad Sci U S A. 2005. PMID: 15932940 Free PMC article.
-
Structure of the human TIP60-C histone exchange and acetyltransferase complex.Nature. 2024 Nov;635(8039):764-769. doi: 10.1038/s41586-024-08011-w. Epub 2024 Sep 11. Nature. 2024. PMID: 39260417 Free PMC article.
-
Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region.EMBO J. 2000 Dec 15;19(24):6814-22. doi: 10.1093/emboj/19.24.6814. EMBO J. 2000. PMID: 11118216 Free PMC article.
References
-
- Belotserkovskaya R. and Berger,S.L. (1999) Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Express., 9, 221–230. - PubMed
-
- Berger S.L., Piña,B., Silverman,N., Marcus,G.A., Agapite,J., Regier,J.L., Triezenberg,S.J. and Guarente,L. (1992) Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell, 70, 251–265. - PubMed
-
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

