Wild-type huntingtin protects from apoptosis upstream of caspase-3
- PMID: 10804212
- PMCID: PMC6772672
- DOI: 10.1523/JNEUROSCI.20-10-03705.2000
Wild-type huntingtin protects from apoptosis upstream of caspase-3
Abstract
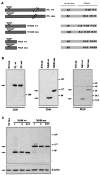
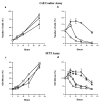
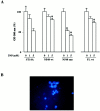
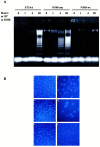
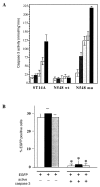
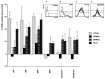
Expansion of a polyglutamine sequence in the N terminus of huntingtin is the gain-of-function event that causes Huntington's disease. This mutation affects primarily the medium-size spiny neurons of the striatum. Huntingtin is expressed in many neuronal and non-neuronal cell types, implying a more general function for the wild-type protein. Here we report that wild-type huntingtin acts by protecting CNS cells from a variety of apoptotic stimuli, including serum withdrawal, death receptors, and pro-apoptotic Bcl-2 homologs. This protection may take place at the level of caspase-9 activation. The full-length protein also modulates the toxicity of the poly-Q expansion. Cells expressing full-length mutant protein are susceptible to fewer death stimuli than cells expressing truncated mutant huntingtin.
Figures

Similar articles
-
Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition.J Neurosci. 1999 Feb 1;19(3):964-73. doi: 10.1523/JNEUROSCI.19-03-00964.1999. J Neurosci. 1999. PMID: 9920660 Free PMC article.
-
Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease.J Neurosci. 2003 Mar 15;23(6):2193-202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. J Neurosci. 2003. PMID: 12657678 Free PMC article.
-
Tissue transglutaminase selectively modifies proteins associated with truncated mutant huntingtin in intact cells.Neurobiol Dis. 2001 Jun;8(3):391-404. doi: 10.1006/nbdi.2001.0390. Neurobiol Dis. 2001. PMID: 11442349
-
The selective vulnerability of nerve cells in Huntington's disease.Neuropathol Appl Neurobiol. 2001 Feb;27(1):1-21. doi: 10.1046/j.0305-1846.2001.00299.x. Neuropathol Appl Neurobiol. 2001. PMID: 11298997 Review.
-
Huntington disease: new insights on the role of huntingtin cleavage.J Neural Transm Suppl. 2000;(58):1-17. doi: 10.1007/978-3-7091-6284-2_1. J Neural Transm Suppl. 2000. PMID: 11128600 Review.
Cited by
-
Preclinical evaluation of stereopure antisense oligonucleotides for allele-selective lowering of mutant HTT.Mol Ther Nucleic Acids. 2024 Jun 11;35(3):102246. doi: 10.1016/j.omtn.2024.102246. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39027419 Free PMC article.
-
Huntingtin promotes cell survival by preventing Pak2 cleavage.J Cell Sci. 2009 Mar 15;122(Pt 6):875-85. doi: 10.1242/jcs.050013. Epub 2009 Feb 24. J Cell Sci. 2009. PMID: 19240112 Free PMC article.
-
Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice.Mol Ther. 2009 Jun;17(6):1053-63. doi: 10.1038/mt.2009.17. Epub 2009 Feb 24. Mol Ther. 2009. PMID: 19240687 Free PMC article.
-
Optimization of trans-Splicing for Huntington's Disease RNA Therapy.Front Neurosci. 2017 Oct 10;11:544. doi: 10.3389/fnins.2017.00544. eCollection 2017. Front Neurosci. 2017. PMID: 29066943 Free PMC article.
-
Reduced cell size, chromosomal aberration and altered proliferation rates are characteristics and confounding factors in the STHdh cell model of Huntington disease.Sci Rep. 2017 Dec 4;7(1):16880. doi: 10.1038/s41598-017-17275-4. Sci Rep. 2017. PMID: 29203806 Free PMC article.
References
-
- Benedetti S, Pirola B, Pollo B, Magrassi L, Bruzzone MG, Rigamonti D, Galli R, Selleri S, Di Meco F, De Fraja C, Vescovi A, Cattaneo E, Finocchiaro G. Gene therapy of experimental brain tumors using neural progenitor cells. Nat Med. 2000;4:447–450. - PubMed
-
- Cattaneo E, Conti L. Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J Neurosci Res. 1998;53:223–234. - PubMed
-
- Cattaneo E, Magrassi L, Santi L, Butti G, Giavazzi A, Pezzotta S. A short term analysis of the behaviour of conditionally immortalized progenitors and primary neuroepithelial cells implanted into the fetal rat brain. Dev Brain Res. 1994;83:197–208. - PubMed
-
- Cattaneo E, De Fraja C, Conti L, Reinach B, Bolis L, Govoni S, Liboi E. Activation of the JAK-STAT pathway leads to proliferation of ST14A CNS progenitor cells. J Biol Chem. 1996a;38:23374–23379. - PubMed
-
- Cattaneo E, Conti L, Gritti A, Frolichsthal P, Govoni S, Vescovi A. Non-virally mediated gene transfer into human Central Nervous System precursor cells. Mol Brain Res. 1996b;42:161–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

