Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends
- PMID: 10793133
- PMCID: PMC14865
- DOI: 10.1091/mbc.11.5.1535
Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends
Abstract
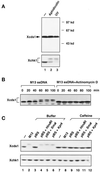
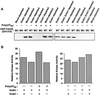
Although homologues of the yeast checkpoint kinases Cds1 and Chk1 have been identified in various systems, the respective roles of these kinases in the responses to damaged and/or unreplicated DNA in vertebrates have not been delineated precisely. Likewise, it is largely unknown how damaged DNA and unreplicated DNA trigger the pathways that contain these effector kinases. We report that Xenopus Cds1 (Xcds1) is phosphorylated and activated by the presence of some simple DNA molecules with double-stranded ends in cell-free Xenopus egg extracts. Xcds1 is not affected by aphidicolin, an agent that induces DNA replication blocks. In contrast, Xenopus Chk1 (Xchk1) responds to DNA replication blocks but not to the presence of double-stranded DNA ends. Immunodepletion of Xcds1 (and/or Xchk1) from egg extracts did not attenuate the cell cycle delay induced by double-stranded DNA ends. These results imply that the cell cycle delay triggered by double-stranded DNA ends either does not involve Xcds1 or uses a factor(s) that can act redundantly with Xcds1.
Figures






Similar articles
-
Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts.Genes Dev. 2000 Nov 1;14(21):2745-56. doi: 10.1101/gad.842500. Genes Dev. 2000. PMID: 11069891 Free PMC article.
-
Xenopus Cds1 is regulated by DNA-dependent protein kinase and ATR during the cell cycle checkpoint response to double-stranded DNA ends.Mol Cell Biol. 2004 Nov;24(22):9968-85. doi: 10.1128/MCB.24.22.9968-9985.2004. Mol Cell Biol. 2004. PMID: 15509799 Free PMC article.
-
The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts.J Cell Biol. 1998 Sep 21;142(6):1559-69. doi: 10.1083/jcb.142.6.1559. J Cell Biol. 1998. PMID: 9744884 Free PMC article.
-
Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways.J Cell Sci. 2000 Nov;113 ( Pt 22)(Pt 22):3889-96. doi: 10.1242/jcs.113.22.3889. J Cell Sci. 2000. PMID: 11058076 Free PMC article. Review.
-
Chk1 in the DNA damage response: conserved roles from yeasts to mammals.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1025-32. doi: 10.1016/j.dnarep.2004.03.003. DNA Repair (Amst). 2004. PMID: 15279789 Review.
Cited by
-
Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts.Genes Dev. 2000 Nov 1;14(21):2745-56. doi: 10.1101/gad.842500. Genes Dev. 2000. PMID: 11069891 Free PMC article.
-
Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation.Curr Biol. 2010 Mar 9;20(5):387-96. doi: 10.1016/j.cub.2010.01.020. Epub 2010 Feb 25. Curr Biol. 2010. PMID: 20188555 Free PMC article.
-
Xenopus Cds1 is regulated by DNA-dependent protein kinase and ATR during the cell cycle checkpoint response to double-stranded DNA ends.Mol Cell Biol. 2004 Nov;24(22):9968-85. doi: 10.1128/MCB.24.22.9968-9985.2004. Mol Cell Biol. 2004. PMID: 15509799 Free PMC article.
-
Identification of dAven, a Drosophila melanogaster ortholog of the cell cycle regulator Aven.Cell Cycle. 2011 Mar 15;10(6):989-98. doi: 10.4161/cc.10.6.15080. Epub 2011 Mar 15. Cell Cycle. 2011. PMID: 21368576 Free PMC article.
-
Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1.Cell Rep. 2013 May 30;3(5):1651-62. doi: 10.1016/j.celrep.2013.04.018. Epub 2013 May 16. Cell Rep. 2013. PMID: 23684611 Free PMC article.
References
-
- Anderson CW. DNA damage and the DNA-activated protein kinase. Trends Biochem Sci. 1993;18:433–437. - PubMed
-
- Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
-
- Blasina A, Van de Weyer I, Laus MC, Luyten WHML, Parker AE, McGowan CH. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1999;9:1–10. - PubMed
-
- Boddy MD, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

