A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5
- PMID: 10779565
- PMCID: PMC25881
- DOI: 10.1073/pnas.090576697
A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5
Abstract
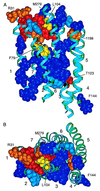
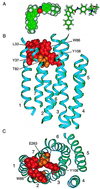
HIV-1 entry into CD4(+) cells requires the sequential interactions of the viral envelope glycoproteins with CD4 and a coreceptor such as the chemokine receptors CCR5 and CXCR4. A plausible approach to blocking this process is to use small molecule antagonists of coreceptor function. One such inhibitor has been described for CCR5: the TAK-779 molecule. To facilitate the further development of entry inhibitors as antiviral drugs, we have explored how TAK-779 acts to prevent HIV-1 infection, and we have mapped its site of interaction with CCR5. We find that TAK-779 inhibits HIV-1 replication at the membrane fusion stage by blocking the interaction of the viral surface glycoprotein gp120 with CCR5. We could identify no amino acid substitutions within the extracellular domain of CCR5 that affected the antiviral action of TAK-779. However, alanine scanning mutagenesis of the transmembrane domains revealed that the binding site for TAK-779 on CCR5 is located near the extracellular surface of the receptor, within a cavity formed between transmembrane helices 1, 2, 3, and 7.
Figures
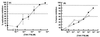
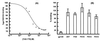

Similar articles
-
Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists.Mol Pharmacol. 2008 Mar;73(3):789-800. doi: 10.1124/mol.107.042101. Epub 2007 Dec 20. Mol Pharmacol. 2008. PMID: 18096812
-
Inhibitory effects of small-molecule CCR5 antagonists on human immunodeficiency virus type 1 envelope-mediated membrane fusion and viral replication.Antimicrob Agents Chemother. 2001 Dec;45(12):3538-43. doi: 10.1128/AAC.45.12.3538-3543.2001. Antimicrob Agents Chemother. 2001. PMID: 11709336 Free PMC article.
-
Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics.Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16249-54. doi: 10.1073/pnas.252469399. Epub 2002 Nov 20. Proc Natl Acad Sci U S A. 2002. PMID: 12444251 Free PMC article.
-
[Viral entry as therapeutic target. Current situation of entry inhibitors].Enferm Infecc Microbiol Clin. 2008 Oct;26 Suppl 11:5-11. doi: 10.1016/s0213-005x(08)76557-1. Enferm Infecc Microbiol Clin. 2008. PMID: 19133215 Review. Spanish.
-
[Chemokine receptors and its importance in the replication cycle of human immunodeficiency virus: clinical and therapeutic implications].Acta Med Port. 2008 Sep-Oct;21(5):497-504. Epub 2009 Jan 16. Acta Med Port. 2008. PMID: 19187693 Review. Portuguese.
Cited by
-
Profile of HIV type 1 coreceptor tropism among Kenyan patients from 2009 to 2010.AIDS Res Hum Retroviruses. 2013 Aug;29(8):1105-9. doi: 10.1089/aid.2012.0284. Epub 2013 May 21. AIDS Res Hum Retroviruses. 2013. PMID: 23617327 Free PMC article.
-
The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C.J Virol. 2003 Apr;77(7):4449-56. doi: 10.1128/jvi.77.7.4449-4456.2003. J Virol. 2003. PMID: 12634405 Free PMC article.
-
Antigenically distinct conformations of CXCR4.J Virol. 2001 Oct;75(19):8957-67. doi: 10.1128/JVI.75.19.8957-8967.2001. J Virol. 2001. PMID: 11533159 Free PMC article.
-
Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors.J Virol. 2004 Apr;78(7):3223-32. doi: 10.1128/jvi.78.7.3223-3232.2004. J Virol. 2004. PMID: 15016843 Free PMC article.
-
Chemokine blockade: a new era in the treatment of rheumatoid arthritis?Arthritis Res Ther. 2004;6(3):93-7. doi: 10.1186/ar1172. Epub 2004 Apr 1. Arthritis Res Ther. 2004. PMID: 15142257 Free PMC article. Review.
References
-
- Fauci A S. N Engl J Med. 1999;341:1046–1050. - PubMed
-
- Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. - PubMed
-
- Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. - PubMed
-
- Lucas G M, Chaisson R E, Moore R D. Ann Intern Med. 1999;131:81–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

