Functional interactions between an atypical NF-kappaB site from the rat CYP2B1 promoter and the transcriptional repressor RBP-Jkappa/CBF1
- PMID: 10773077
- PMCID: PMC105370
- DOI: 10.1093/nar/28.10.2091
Functional interactions between an atypical NF-kappaB site from the rat CYP2B1 promoter and the transcriptional repressor RBP-Jkappa/CBF1
Abstract
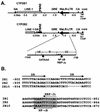
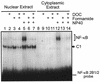
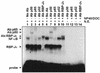
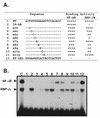
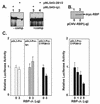
The phenobarbital-inducible rat cytochrome P450 (CYP) 2B1 and 2B2 proteins are encoded by homologous genes whose promoters contain a mammalian-apparent long terminal repeat retrotransposon (MaLR). An NF-kappaB-like site within the MaLR forms multiple protein-DNA complexes with rat liver and HeLa cell nuclear extracts. Using antibody supershift assays, we have identified these complexes as NF-kappaB and RPB-Jkappa/CBF1. Competition assays using a series of single site mutant oligonucleotides reveal that the recognition sites for these two factors overlap. We also show that the CYP2B1/2 NF-kappaB element, but not the Igkappa NF-kappaB element, can repress transcription in vitro when positioned upstream of the heterologous adenovirus major late core promoter. In addition, RBP-Jkappa over-expressed in COS-7 cells repressed expression in vivo from an SV40-luciferase reporter construct that contained the CYP2B1/2 NF-kappaB element. Finally, we observe similar levels of NF-kappaB and RBP-Jkappa binding activities in nuclear extracts prepared from control and phenobarbital-induced rat livers. The results suggest that RBP-Jkappa/CBF1 binds an atypical NF-kappaB site in the CYP2B1/2 promoters and may help to maintain a low level of expression in the absence of inducer.
Figures
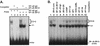

Similar articles
-
Purification of a novel MHC class I element binding activity from thymus nuclear extracts reveals that thymic RBP-Jkappa/CBF1 binds to NF-kappaB-like elements.J Immunol. 1996 Jun 15;156(12):4672-9. J Immunol. 1996. PMID: 8648111
-
The Ku antigen-recombination signal-binding protein Jkappa complex binds to the nuclear factor-kappaB p50 promoter and acts as a positive regulator of p50 expression in human gastric cancer cells.J Biol Chem. 2004 Jan 2;279(1):231-7. doi: 10.1074/jbc.M308609200. Epub 2003 Oct 21. J Biol Chem. 2004. PMID: 14570916
-
Nuclear factor-1 motif and redundant regulatory elements comprise phenobarbital-responsive enhancer in CYP2B1/2.DNA Cell Biol. 1998 May;17(5):461-70. doi: 10.1089/dna.1998.17.461. DNA Cell Biol. 1998. PMID: 9628589
-
NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa.Mol Cell Biol. 1998 Apr;18(4):2077-88. doi: 10.1128/MCB.18.4.2077. Mol Cell Biol. 1998. PMID: 9528780 Free PMC article.
-
Binding of C/EBP and RBP (CBF1) to overlapping sites regulates interleukin-6 gene expression.J Biol Chem. 2002 Nov 8;277(45):42438-46. doi: 10.1074/jbc.M207363200. Epub 2002 Aug 27. J Biol Chem. 2002. PMID: 12200447
Cited by
-
The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis.Mol Cell Biol. 2008 Oct;28(20):6402-12. doi: 10.1128/MCB.00299-08. Epub 2008 Aug 18. Mol Cell Biol. 2008. PMID: 18710934 Free PMC article.
-
A novel RBP-J kappa-dependent switch from C/EBP beta to C/EBP zeta at the C/EBP binding site on the C-reactive protein promoter.J Immunol. 2007 Jun 1;178(11):7302-9. doi: 10.4049/jimmunol.178.11.7302. J Immunol. 2007. PMID: 17513780 Free PMC article.
-
NF-kappaB serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator.J Virol. 2009 May;83(9):4435-46. doi: 10.1128/JVI.01999-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244329 Free PMC article.
-
The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses.J Virol. 2015 Mar;89(5):2575-89. doi: 10.1128/JVI.02791-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520508 Free PMC article.
-
Loss of the Notch effector RBPJ promotes tumorigenesis.J Exp Med. 2015 Jan 12;212(1):37-52. doi: 10.1084/jem.20121192. Epub 2014 Dec 15. J Exp Med. 2015. PMID: 25512468 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

