Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes
- PMID: 10756049
- PMCID: PMC111951
- DOI: 10.1128/jvi.74.9.4335-4350.2000
Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes
Abstract
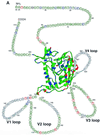
To elucidate the evolutionary mechanisms of the human immunodeficiency virus type 1 gp120 envelope glycoprotein at the single-site level, the degree of amino acid variation and the numbers of synonymous and nonsynonymous substitutions were examined in 186 nucleotide sequences for gp120 (subtype B). Analyses of amino acid variabilities showed that the level of variability was very different from site to site in both conserved (C1 to C5) and variable (V1 to V5) regions previously assigned. To examine the relative importance of positive and negative selection for each amino acid position, the numbers of synonymous and nonsynonymous substitutions that occurred at each codon position were estimated by taking phylogenetic relationships into account. Among the 414 codon positions examined, we identified 33 positions where nonsynonymous substitutions were significantly predominant. These positions where positive selection may be operating, which we call putative positive selection (PS) sites, were found not only in the variable loops but also in the conserved regions (C1 to C4). In particular, we found seven PS sites at the surface positions of the alpha-helix (positions 335 to 347 in the C3 region) in the opposite face for CD4 binding. Furthermore, two PS sites in the C2 region and four PS sites in the C4 region were detected in the same face of the protein. The PS sites found in the C2, C3, and C4 regions were separated in the amino acid sequence but close together in the three-dimensional structure. This observation suggests the existence of discontinuous epitopes in the protein's surface including this alpha-helix, although the antigenicity of this area has not been reported yet.
Figures





Similar articles
-
[Sequence variation of HIV and bioinformatics].Uirusu. 2004 Jun;54(1):33-8. doi: 10.2222/jsv.54.33. Uirusu. 2004. PMID: 15449902 Review. Japanese.
-
The effect of low-profile serine substitutions in the V3 loop of HIV-1 gp120 IIIB/LAI on the immunogenicity of the envelope protein.Virology. 1998 Nov 10;251(1):59-70. doi: 10.1006/viro.1998.9392. Virology. 1998. PMID: 9813203
-
Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop.J Virol. 2002 Apr;76(8):3852-64. doi: 10.1128/jvi.76.8.3852-3864.2002. J Virol. 2002. PMID: 11907225 Free PMC article.
-
Presence of multiple genetic subtypes of human immunodeficiency virus type 1 proviruses in Uganda.AIDS Res Hum Retroviruses. 1994 Nov;10(11):1543-50. doi: 10.1089/aid.1994.10.1543. AIDS Res Hum Retroviruses. 1994. PMID: 7888209
-
Combinatorial libraries, epitope structure and the prediction of protein conformations.Immunol Today. 1997 Mar;18(3):108-10. doi: 10.1016/s0167-5699(97)01024-4. Immunol Today. 1997. PMID: 9078681 Review. No abstract available.
Cited by
-
The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia.Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15160-5. doi: 10.1073/pnas.0605513103. Epub 2006 Oct 2. Proc Natl Acad Sci U S A. 2006. PMID: 17015824 Free PMC article.
-
Molecular evolution of the hepatitis delta virus antigen gene: recombination or positive selection?J Mol Evol. 2004 Dec;59(6):815-26. doi: 10.1007/s00239-004-0112-x. J Mol Evol. 2004. PMID: 15599513
-
Molecular characterization of HIV-1 subtype C gp-120 regions potentially involved in virus adaptive mechanisms.PLoS One. 2014 Apr 30;9(4):e95183. doi: 10.1371/journal.pone.0095183. eCollection 2014. PLoS One. 2014. PMID: 24788065 Free PMC article.
-
Identification of Owl Monkey CD4 Receptors Broadly Compatible with Early-Stage HIV-1 Isolates.J Virol. 2015 Aug;89(16):8611-22. doi: 10.1128/JVI.00890-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063421 Free PMC article.
-
Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes.J Virol. 2004 Feb;78(4):1962-70. doi: 10.1128/jvi.78.4.1962-1970.2004. J Virol. 2004. PMID: 14747561 Free PMC article.
References
-
- Adachi J, Hasegawa M. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs vol. 28. Tokyo: Institute of Statistical Mathematics; 1996. pp. 1–150.
-
- Bonhoeffer S, Holmes E C, Nowak M A. Cause of HIV diversity. Nature. 1995;376:125. - PubMed
-
- Bush R M, Fitch W M, Bender C A, Cox N J. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol. 1999;16:1457–1465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

