Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria
- PMID: 10713172
- PMCID: PMC85448
- DOI: 10.1128/MCB.20.7.2488-2497.2000
Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria
Abstract
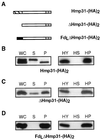
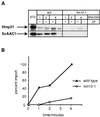
A number of microaerophilic eukaryotes lack mitochondria but possess another organelle involved in energy metabolism, the hydrogenosome. Limited phylogenetic analyses of nuclear genes support a common origin for these two organelles. We have identified a protein of the mitochondrial carrier family in the hydrogenosome of Trichomonas vaginalis and have shown that this protein, Hmp31, is phylogenetically related to the mitochondrial ADP-ATP carrier (AAC). We demonstrate that the hydrogenosomal AAC can be targeted to the inner membrane of mitochondria isolated from Saccharomyces cerevisiae through the Tim9-Tim10 import pathway used for the assembly of mitochondrial carrier proteins. Conversely, yeast mitochondrial AAC can be targeted into the membranes of hydrogenosomes. The hydrogenosomal AAC contains a cleavable, N-terminal presequence; however, this sequence is not necessary for targeting the protein to the organelle. These data indicate that the membrane-targeting signal(s) for hydrogenosomal AAC is internal, similar to that found for mitochondrial carrier proteins. Our findings indicate that the membrane carriers and membrane protein-targeting machinery of hydrogenosomes and mitochondria have a common evolutionary origin. Together, they provide strong evidence that a single endosymbiont evolved into a progenitor organelle in early eukaryotic cells that ultimately give rise to these two distinct organelles and support the hydrogen hypothesis for the origin of the eukaryotic cell.
Figures





Similar articles
-
Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases.Eukaryot Cell. 2008 Oct;7(10):1750-7. doi: 10.1128/EC.00206-08. Epub 2008 Aug 1. Eukaryot Cell. 2008. PMID: 18676956 Free PMC article.
-
The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis.PLoS One. 2011;6(9):e24428. doi: 10.1371/journal.pone.0024428. Epub 2011 Sep 15. PLoS One. 2011. PMID: 21935410 Free PMC article.
-
A divergent ADP/ATP carrier in the hydrogenosomes of Trichomonas gallinae argues for an independent origin of these organelles.Mol Microbiol. 2004 Mar;51(5):1439-46. doi: 10.1111/j.1365-2958.2004.03918.x. Mol Microbiol. 2004. PMID: 14982636
-
Biogenesis of the hydrogenosome in the anaerobic protist Trichomonas vaginalis.J Parasitol. 1993 Oct;79(5):664-70. J Parasitol. 1993. PMID: 8410536 Review.
-
The ADP and ATP transport in mitochondria and its carrier.Biochim Biophys Acta. 2008 Oct;1778(10):1978-2021. doi: 10.1016/j.bbamem.2008.04.011. Epub 2008 May 2. Biochim Biophys Acta. 2008. PMID: 18510943 Review.
Cited by
-
Novel variants of human SCaMC-3, an isoform of the ATP-Mg/P(i) mitochondrial carrier, generated by alternative splicing from 3'-flanking transposable elements.Biochem J. 2005 Aug 1;389(Pt 3):647-55. doi: 10.1042/BJ20050283. Biochem J. 2005. PMID: 15801905 Free PMC article.
-
Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression.Mol Cell Proteomics. 2010 Jul;9(7):1554-66. doi: 10.1074/mcp.M000022-MCP201. Epub 2010 May 13. Mol Cell Proteomics. 2010. PMID: 20467041 Free PMC article.
-
Role of YHM1, encoding a mitochondrial carrier protein, in iron distribution of yeast.Biochem J. 2004 Mar 1;378(Pt 2):599-607. doi: 10.1042/BJ20031387. Biochem J. 2004. PMID: 14629196 Free PMC article.
-
Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases.Eukaryot Cell. 2008 Oct;7(10):1750-7. doi: 10.1128/EC.00206-08. Epub 2008 Aug 1. Eukaryot Cell. 2008. PMID: 18676956 Free PMC article.
-
Minimization of extracellular space as a driving force in prokaryote association and the origin of eukaryotes.Biol Direct. 2014 Nov 18;9(1):24. doi: 10.1186/1745-6150-9-24. Biol Direct. 2014. PMID: 25406691 Free PMC article.
References
-
- Andersson S G E, Kurland C G. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol. 1999;2:535–541. - PubMed
-
- Andersson S G E, Zomorodipour A, Andersson J P, Sicheritz-Ponten T, Alsmark U C M, Podowski R M, Naslund A K, Eriksson A-S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
