The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577
- PMID: 10708456
- PMCID: PMC111840
- DOI: 10.1128/jvi.74.7.3388-3398.2000
The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577
Abstract
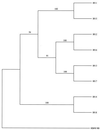
The primary sequence of the long unique region L-DNA (L for low GC) of rhesus monkey rhadinovirus (RRV) isolate 26-95 was determined. The L-DNA consists of 130,733 bp that contain 84 open reading frames (ORFs). The overall organization of the RRV26-95 genome was found to be very similar to that of human Kaposi sarcoma-associated herpesvirus (KSHV). BLAST search analysis revealed that in almost all cases RRV26-95 coding sequences have a greater degree of similarity to corresponding KSHV sequences than to other herpesviruses. All of the ORFs present in KSHV have at least one homologue in RRV26-95 except K3 and K5 (bovine herpesvirus-4 immediate-early protein homologues), K7 (nut-1), and K12 (Kaposin). RRV26-95 contains one MIP-1 and eight interferon regulatory factor (vIRF) homologues compared to three MIP-1 and four vIRF homologues in KSHV. All homologues are correspondingly located in KSHV and RRV with the exception of dihydrofolate reductase (DHFR). DHFR is correspondingly located near the left end of the genome in RRV26-95 and herpesvirus saimiri (HVS), but in KSHV the DHFR gene is displaced 16,069 nucleotides in a rightward direction in the genome. DHFR is also unusual in that the RRV26-95 DHFR more closely resembles HVS DHFR (74% similarity) than KSHV DHFR (55% similarity). Of the 84 ORFs in RRV26-95, 83 contain sequences similar to the recently determined sequences of the independent RRV isolate 17577. RRV26-95 and RRV17577 sequences differ in that ORF 67.5 sequences contained in RRV26-95 were not found in RRV17577. In addition, ORF 4 is significantly shorter in RRV26-95 than was reported for RRV17577 (395 versus 645 amino acids). Only four of the corresponding ORFs between RRV26-95 and RRV17577 exhibited less than 95% sequence identity: glycoproteins H and L, uracil DNA glucosidase, and a tegument protein (ORF 67). Both RRV26-95 and RRV17577 have unique ORFs between positions 21444 to 21752 and 110910 to 114899 in a rightward direction and from positions 116524 to 111082 in a leftward direction that are not found in KSHV. Our analysis indicates that RRV26-95 and RRV17577 are clearly independent isolates of the same virus species and that both are closely related in structural organization and overall sequence to KSHV. The availability of detailed sequence information, the ability to grow RRV lytically in cell culture, and the ability to infect monkeys experimentally with RRV will facilitate the construction of mutant strains of virus for evaluating the contribution of individual genes to biological properties.
Figures




Similar articles
-
Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus.J Virol. 2015 Apr;89(7):3888-909. doi: 10.1128/JVI.03597-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609822 Free PMC article.
-
Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8.J Virol. 1999 Apr;73(4):3040-53. doi: 10.1128/JVI.73.4.3040-3053.1999. J Virol. 1999. PMID: 10074154 Free PMC article.
-
Construction of an infectious rhesus rhadinovirus bacterial artificial chromosome for the analysis of Kaposi's sarcoma-associated herpesvirus-related disease development.J Virol. 2007 Mar;81(6):2957-69. doi: 10.1128/JVI.01997-06. Epub 2007 Jan 10. J Virol. 2007. PMID: 17215283 Free PMC article.
-
Rhesus monkey rhadinovirus: a model for the study of KSHV.Curr Top Microbiol Immunol. 2007;312:43-69. doi: 10.1007/978-3-540-34344-8_2. Curr Top Microbiol Immunol. 2007. PMID: 17089793 Review.
-
Rhesus macaque rhadinovirus-associated disease.Curr Opin Virol. 2013 Jun;3(3):245-50. doi: 10.1016/j.coviro.2013.05.016. Epub 2013 Jun 6. Curr Opin Virol. 2013. PMID: 23747119 Free PMC article. Review.
Cited by
-
Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus.J Virol. 2015 Apr;89(7):3888-909. doi: 10.1128/JVI.03597-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609822 Free PMC article.
-
The collagen repeat sequence is a determinant of the degree of herpesvirus saimiri STP transforming activity.J Virol. 2000 Sep;74(17):8102-10. doi: 10.1128/jvi.74.17.8102-8110.2000. J Virol. 2000. PMID: 10933720 Free PMC article.
-
Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase.J Virol. 2001 Mar;75(5):2345-52. doi: 10.1128/JVI.75.5.2345-2352.2001. J Virol. 2001. PMID: 11160738 Free PMC article.
-
Eph receptors: the bridge linking host and virus.Cell Mol Life Sci. 2020 Jun;77(12):2355-2365. doi: 10.1007/s00018-019-03409-6. Epub 2019 Dec 31. Cell Mol Life Sci. 2020. PMID: 31893311 Free PMC article. Review.
-
A critical Sp1 element in the rhesus rhadinovirus (RRV) Rta promoter confers high-level activity that correlates with cellular permissivity for viral replication.Virology. 2014 Jan 5;448:196-209. doi: 10.1016/j.virol.2013.10.013. Epub 2013 Oct 29. Virology. 2014. PMID: 24314650 Free PMC article.
References
-
- Alexander L, Lee H, Rosenzweig M, Jung J U, Desrosiers R C. An EGFP-containing vector system that facilitates stable and transient expression assays. BioTechniques. 1997;23:64–66. - PubMed
-
- Blake N W, Moghaddam A, Rao P, Kaur A, Glickman R, Cho Y, Marchini A, Haigh T, Johnson R P, Rickinson A B, Wang F. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J Virol. 1999;73:7381–7389. - PMC - PubMed
-
- Boshoff C, Whitby D, Hatzioannou T, Fisher C, van der Walt J, Hatzakis A, Weiss R A, Schultz T F. Kaposi's sarcoma associated herpesvirus in HIV-negative Kaposi sarcoma. Lancet. 1995;345:1043–1044. - PubMed
-
- Brakenhoff J P, de Hon F D, Fontaine V, ten Boekel E, Schooltink H, Rose-John S, Heinrich P C, Content J, Aarden L A. Development of a human interleukin-6 receptor antagonist. J Biol Chem. 1994;269:86–93. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

