Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa
- PMID: 10692351
- PMCID: PMC94443
- DOI: 10.1128/JB.182.6.1481-1491.2000
Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa
Abstract
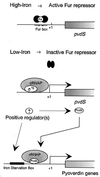
In Pseudomonas aeruginosa, iron modulates gene expression through a cascade of negative and positive regulatory proteins. The master regulator Fur is involved in iron-dependent repression of several genes. One of these genes, pvdS, was predicted to encode a putative sigma factor responsible for the transcription of a subset of genes of the Fur regulon. PvdS appears to belong to a structurally and functionally distinct subgroup of the extracytoplasmic function family of alternative sigma factors. Members of this subgroup, also including PbrA from Pseudomonas fluorescens, PfrI and PupI from Pseudomonas putida, and FecI from Escherichia coli, are controlled by the Fur repressor, and they activate transcription of genes for the biosynthesis or the uptake of siderophores. Evidence is provided that the PvdS protein of P. aeruginosa is endowed with biochemical properties of eubacterial sigma factors, as it spontaneously forms 1:1 complexes with the core fraction of RNA polymerase (RNAP, alpha(2)betabeta' subunits), thereby promoting in vitro binding of the PvdS-RNAP holoenzyme to the promoter region of the pvdA gene. These functional features of PvdS are consistent with the presence of structural domains predicted to be involved in core RNAP binding, promoter recognition, and open complex formation. The activity of pyoverdin biosynthetic (pvd) promoters was significantly lower in E. coli overexpressing the multicopy pvdS gene than in wild-type P. aeruginosa PAO1 carrying the single gene copy, and pvd::lacZ transcriptional fusions were silent in both pfrI (the pvdS homologue) and pfrA (a positive regulator of pseudobactin biosynthetic genes) mutants of P. putida WCS358, while they are expressed at PAO1 levels in wild-type WCS358. Moreover, the PvdS-RNAP holoenzyme purified from E. coli lacked the ability to generate in vitro transcripts from the pvdA promoter. These observations suggest that at least one additional positive regulator could be required for full activity of the PvdS-dependent transcription complex both in vivo and in vitro. This is consistent with the presence of a putative activator binding site (the iron starvation box) at variable distance from the transcription initiation sites of promoters controlled by the iron starvation sigma factors PvdS, PfrI, and PbrA of fluorescent pseudomonads.
Figures




Similar articles
-
Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity.J Bacteriol. 1996 Apr;178(8):2299-313. doi: 10.1128/jb.178.8.2299-2313.1996. J Bacteriol. 1996. PMID: 8636031 Free PMC article.
-
A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO.Mol Gen Genet. 1995 Jul 22;248(1):17-24. doi: 10.1007/BF02456609. Mol Gen Genet. 1995. PMID: 7651323
-
Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa.J Bacteriol. 2001 Mar;183(6):2151-5. doi: 10.1128/JB.183.6.2151-2155.2001. J Bacteriol. 2001. PMID: 11222621 Free PMC article.
-
How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events.Biometals. 2007 Jun;20(3-4):587-601. doi: 10.1007/s10534-006-9067-2. Epub 2006 Dec 22. Biometals. 2007. PMID: 17186376 Review.
-
Iron uptake regulation in Pseudomonas aeruginosa.Biometals. 2009 Feb;22(1):15-22. doi: 10.1007/s10534-008-9193-0. Epub 2009 Jan 8. Biometals. 2009. PMID: 19130263 Review.
Cited by
-
Genome Sequence of Azospirillum brasilense CBG497 and Comparative Analyses of Azospirillum Core and Accessory Genomes provide Insight into Niche Adaptation.Genes (Basel). 2012 Sep 28;3(4):576-602. doi: 10.3390/genes3040576. Genes (Basel). 2012. PMID: 24705077 Free PMC article.
-
High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections.BMC Microbiol. 2017 Feb 3;17(1):30. doi: 10.1186/s12866-017-0941-6. BMC Microbiol. 2017. PMID: 28158967 Free PMC article.
-
Phage-mediated resolution of genetic conflict alters the evolutionary trajectory of Pseudomonas aeruginosa lysogens.mSystems. 2024 Sep 17;9(9):e0080124. doi: 10.1128/msystems.00801-24. Epub 2024 Aug 21. mSystems. 2024. PMID: 39166874 Free PMC article.
-
Involvement of AlgQ in transcriptional regulation of pyoverdine genes in Pseudomonas aeruginosa PAO1.J Bacteriol. 2005 Aug;187(15):5097-107. doi: 10.1128/JB.187.15.5097-5107.2005. J Bacteriol. 2005. PMID: 16030202 Free PMC article.
-
A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa.PLoS One. 2013 Dec 30;8(12):e84775. doi: 10.1371/journal.pone.0084775. eCollection 2013. PLoS One. 2013. PMID: 24386415 Free PMC article.
References
-
- Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmatic stimuli. Mol Microbiol. 1995;18:163–174. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Brutsche S, Braun V. SigX of Bacillus subtilis replaces the ECF sigma factor FecI of Escherichia coli and is inhibited by RsiX. Mol Gen Genet. 1997;256:416–425. - PubMed
-
- Callanan M, Sexton R, Dowling D N, O'Gara F. Regulation of the iron uptake genes in Pseudomonas fluorescens M114 by pseudobactin M114: the pbrA sigma factor does not mediate the siderophore regulatory response. FEMS Microbiol Lett. 1996;144:61–66. - PubMed
-
- Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

