Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10)
- PMID: 10677520
- PMCID: PMC26498
- DOI: 10.1073/pnas.97.4.1695
Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10)
Abstract
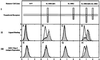
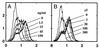
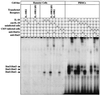
We identified a viral IL-10 homolog encoded by an ORF (UL111a) within the human cytomegalovirus (CMV) genome, which we designated cmvIL-10. cmvIL-10 can bind to the human IL-10 receptor and can compete with human IL-10 for binding sites, despite the fact that these two proteins are only 27% identical. cmvIL-10 requires both subunits of the IL-10 receptor complex to induce signal transduction events and biological activities. The structure of the cmvIL-10 gene is unique by itself. The gene retained two of four introns of the IL-10 gene, but the length of the introns was reduced. We demonstrated that cmvIL-10 is expressed in CMV-infected cells. Thus, expression of cmvIL-10 extends the range of counter measures developed by CMV to circumvent detection and destruction by the host immune system.
Figures




Similar articles
-
Human Cytomegalovirus-Encoded Human Interleukin-10 (IL-10) Homolog Amplifies Its Immunomodulatory Potential by Upregulating Human IL-10 in Monocytes.J Virol. 2016 Mar 28;90(8):3819-3827. doi: 10.1128/JVI.03066-15. Print 2016 Apr. J Virol. 2016. PMID: 26792743 Free PMC article.
-
Human Cytomegalovirus UL111A and US27 Gene Products Enhance the CXCL12/CXCR4 Signaling Axis via Distinct Mechanisms.J Virol. 2018 Feb 12;92(5):e01981-17. doi: 10.1128/JVI.01981-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237840 Free PMC article.
-
Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1.Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9404-9. doi: 10.1073/pnas.152147499. Epub 2002 Jul 1. Proc Natl Acad Sci U S A. 2002. PMID: 12093920 Free PMC article.
-
Human Cytomegalovirus Interleukin 10 Homologs: Facing the Immune System.Front Cell Infect Microbiol. 2020 Jun 9;10:245. doi: 10.3389/fcimb.2020.00245. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32582563 Free PMC article. Review.
-
Activation and regulation of human cytomegalovirus early genes.Intervirology. 1996;39(5-6):361-77. doi: 10.1159/000150507. Intervirology. 1996. PMID: 9130046 Review.
Cited by
-
Is human cytomegalovirus a target in cancer therapy?Oncotarget. 2011 Dec;2(12):1329-38. doi: 10.18632/oncotarget.383. Oncotarget. 2011. PMID: 22246171 Free PMC article. Review.
-
IL-27 expression regulation and its effects on adaptive immunity against viruses.Front Immunol. 2024 Jun 20;15:1395921. doi: 10.3389/fimmu.2024.1395921. eCollection 2024. Front Immunol. 2024. PMID: 38966644 Free PMC article. Review.
-
Involvement of Salmonella pathogenicity island 2 in the up-regulation of interleukin-10 expression in macrophages: role of protein kinase A signal pathway.Infect Immun. 2004 Apr;72(4):1964-73. doi: 10.1128/IAI.72.4.1964-1973.2004. Infect Immun. 2004. PMID: 15039316 Free PMC article.
-
Cell type-specific regulation of IL-10 expression in inflammation and disease.Immunol Res. 2010 Jul;47(1-3):185-206. doi: 10.1007/s12026-009-8150-5. Immunol Res. 2010. PMID: 20087682 Free PMC article. Review.
-
A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection.J Virol. 2004 Feb;78(3):1440-7. doi: 10.1128/jvi.78.3.1440-1447.2004. J Virol. 2004. PMID: 14722299 Free PMC article.
References
-
- Moore K W, O'Garra A, de Waal M, Vieira P, Mosmann T R. Annu Rev Immunol. 1993;11:165–190. - PubMed
-
- Fossa S D, Aamdal S, Naume B, Gallati H. Acta Oncol. 1995;34:599–603. - PubMed
-
- Mori N, Prager D. Leuk Lymphoma. 1998;29:239–248. - PubMed
-
- Halak B K, Maguire H C, Lattime E C. Cancer Res. 1999;59:911–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

