A family of chromatin remodeling factors related to Williams syndrome transcription factor
- PMID: 10655480
- PMCID: PMC15513
- DOI: 10.1073/pnas.97.3.1038
A family of chromatin remodeling factors related to Williams syndrome transcription factor
Abstract
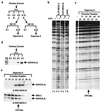
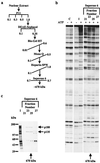
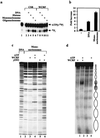
Chromatin remodeling complexes have been implicated in the disruption or reformation of nucleosomal arrays resulting in modulation of transcription, DNA replication, and DNA repair. Here we report the isolation of WCRF, a new chromatin-remodeling complex from HeLa cells. WCRF is composed of two subunits, WCRF135, the human homolog of Drosophila ISWI, and WCRF180, a protein related to the Williams syndrome transcription factor. WCRF180 is a member of a family of proteins sharing a putative heterochromatin localization domain, a PHD finger, and a bromodomain, prevalent in factors involved in regulation of chromatin structure.
Figures

Similar articles
-
Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits.Mol Cell. 1999 Feb;3(2):247-53. doi: 10.1016/s1097-2765(00)80315-9. Mol Cell. 1999. PMID: 10078207
-
WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci.EMBO J. 2002 May 1;21(9):2231-41. doi: 10.1093/emboj/21.9.2231. EMBO J. 2002. PMID: 11980720 Free PMC article.
-
ISWI is an ATP-dependent nucleosome remodeling factor.Mol Cell. 1999 Feb;3(2):239-45. doi: 10.1016/s1097-2765(00)80314-7. Mol Cell. 1999. PMID: 10078206
-
The mammalian SWI/SNF complex and the control of cell growth.Semin Cell Dev Biol. 1999 Apr;10(2):189-95. doi: 10.1006/scdb.1999.0300. Semin Cell Dev Biol. 1999. PMID: 10441072 Review.
-
Transcriptional regulation: SWItching circuitry.Curr Biol. 1999 Mar 25;9(6):R221-4. doi: 10.1016/s0960-9822(99)80134-1. Curr Biol. 1999. PMID: 10209086 Review.
Cited by
-
Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription.Genes Dev. 2001 Nov 1;15(21):2837-51. doi: 10.1101/gad.937401. Genes Dev. 2001. PMID: 11691835 Free PMC article.
-
CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality.Nucleic Acids Res. 2017 Oct 13;45(18):10534-10554. doi: 10.1093/nar/gkx711. Nucleic Acids Res. 2017. PMID: 28977666 Free PMC article.
-
MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response.Cell Rep. 2012 Dec 27;2(6):1657-69. doi: 10.1016/j.celrep.2012.11.018. Cell Rep. 2012. PMID: 23260667 Free PMC article.
-
ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B.Mol Biol Cell. 2002 Jan;13(1):25-39. doi: 10.1091/mbc.01-09-0441. Mol Biol Cell. 2002. PMID: 11809820 Free PMC article.
-
Pathogenic variants in SMARCA5, a chromatin remodeler, cause a range of syndromic neurodevelopmental features.Sci Adv. 2021 May 12;7(20):eabf2066. doi: 10.1126/sciadv.abf2066. Print 2021 May. Sci Adv. 2021. PMID: 33980485 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

