A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes
- PMID: 10655476
- PMCID: PMC15505
- DOI: 10.1073/pnas.97.3.1014
A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes
Abstract
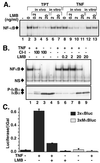
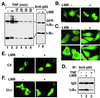
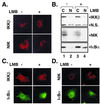
Appropriate subcellular localization is crucial for regulation of NF-kappaB function. Herein, we show that latent NF-kappaB complexes can enter and exit the nucleus in preinduction states. The nuclear export inhibitor leptomycin B (LMB) sequestered NF-kappaB/IkappaBalpha complexes in the nucleus. Using deletion and site-directed mutagenesis, we identified a previously uncharacterized nuclear export sequence in residues 45-54 of IkappaBalpha that was required for cytoplasmic localization of inactive complexes. This nuclear export sequence also caused nuclear exclusion of heterologous proteins in a LMB-sensitive manner. Importantly, a LMB-insensitive CRM1 mutant (Crm1-K1) abolished LMB-induced nuclear accumulation of the inactive complexes. Moreover, a cell-permeable p50 NF-kappaB nuclear localization signal peptide also blocked these LMB effects. These results suggest that NF-kappaB/IkappaBalpha complexes shuttle between the cytoplasm and nucleus by a nuclear localization signal-dependent nuclear import and a CRM1-dependent nuclear export. The LMB-induced nuclear complexes could not bind DNA and were inaccessible to signaling events, because LMB inhibited NF-kappaB activation without affecting the subcellular localization of upstream kinases IKKbeta and NIK. Our findings indicate that the dominant nuclear export over nuclear import contributes to the largely cytoplasmic localization of the inactive complexes to achieve efficient NF-kappaB activation by extracellular signals.
Figures



Similar articles
-
IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2.J Biol Chem. 2000 Nov 17;275(46):36441-9. doi: 10.1074/jbc.M004751200. J Biol Chem. 2000. PMID: 10969074
-
Regulation of RelA subcellular localization by a putative nuclear export signal and p50.Mol Cell Biol. 1999 Oct;19(10):7088-95. doi: 10.1128/MCB.19.10.7088. Mol Cell Biol. 1999. PMID: 10490645 Free PMC article.
-
Evidence for a role of CRM1 in signal-mediated nuclear protein export.Science. 1997 Oct 3;278(5335):141-4. doi: 10.1126/science.278.5335.141. Science. 1997. PMID: 9311922
-
Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs.Curr Med Chem. 2003 May;10(9):741-8. doi: 10.2174/0929867033457791. Curr Med Chem. 2003. PMID: 12678777 Review.
-
Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export.Ann N Y Acad Sci. 1999;886:23-36. doi: 10.1111/j.1749-6632.1999.tb09397.x. Ann N Y Acad Sci. 1999. PMID: 10667200 Review.
Cited by
-
Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFα/NF-κB response.Proc Natl Acad Sci U S A. 2016 Apr 19;113(16):4326-31. doi: 10.1073/pnas.1522372113. Epub 2016 Apr 5. Proc Natl Acad Sci U S A. 2016. PMID: 27051065 Free PMC article.
-
Bacterial interference with canonical NFκB signalling.Microbiology (Reading). 2013 Oct;159(Pt 10):2001-2013. doi: 10.1099/mic.0.069369-0. Epub 2013 Jul 19. Microbiology (Reading). 2013. PMID: 23873783 Free PMC article. Review.
-
CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications.Leukemia. 2014 Jan;28(1):155-65. doi: 10.1038/leu.2013.115. Epub 2013 Apr 16. Leukemia. 2014. PMID: 23588715 Free PMC article.
-
A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11199-204. doi: 10.1073/pnas.172383099. Epub 2002 Aug 8. Proc Natl Acad Sci U S A. 2002. PMID: 12169669 Free PMC article.
-
Exportin-1-Dependent Nuclear Export of DEAD-box Helicase DDX3X is Central to its Role in Antiviral Immunity.Cells. 2019 Sep 30;8(10):1181. doi: 10.3390/cells8101181. Cells. 2019. PMID: 31575075 Free PMC article.
References
-
- Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. - PubMed
-
- Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. - PubMed
-
- Baeuerle P A, Baltimore D. Cell. 1988;53:211–217. - PubMed
-
- Arenzana-Seisdedos F, Turpin T, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. J Cell Sci. 1997;110:369–378. - PubMed
-
- Cheng Q, Cant C A, Moll T, Hofer-Warbinek R, Wagner E, Birnstiel M L, Bach F H, de Martin R. J Biol Chem. 1994;269:13551–13557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

