Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system
- PMID: 10655350
- PMCID: PMC86153
- DOI: 10.1128/JCM.38.2.586-590.2000
Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system
Abstract
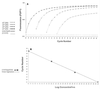
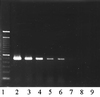
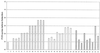
The Light Cycler technique combines rapid in vitro amplification of DNA in glass capillaries with real-time species determination and quantification of DNA load. We have established a quantitative PCR protocol for two clinically important pathogens, Candida albicans and Aspergillus fumigatus. The sensitivity of the assay was comparable to those of previously described PCR protocols (5 CFU/ml). Specific detection of C. albicans and A. fumigatus could be achieved. The assay showed a high reproducibility of 96 to 99%. The assay was linear in a range between 10(1) and 10(4) Aspergillus conidia. As capillaries do not have to be reopened for post-PCR analysis, the risk of carryover contaminations could be minimized. The Light Cycler allowed quantification of the fungal loads in a limited number of clinical specimens from patients with hematological malignancies and histologically proven invasive fungal infections. Five of nine positive samples had fungal loads between 5 and 10 CFU/ml of blood, two of nine positive samples had fungal loads between 10 and 100 CFU/ml of blood, and two of nine samples had fungal loads of more than 100 CFU/ml of blood. All samples were also found to be PCR positive by PCR-enzyme-linked immunosorbent assay analysis.
Figures

Similar articles
-
Monochrome LightCycler PCR assay for detection and quantification of five common species of Candida and Aspergillus.J Med Microbiol. 2005 Mar;54(Pt 3):243-248. doi: 10.1099/jmm.0.45856-0. J Med Microbiol. 2005. PMID: 15713607
-
Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients.J Clin Microbiol. 2003 May;41(5):1811-8. doi: 10.1128/JCM.41.5.1811-1818.2003. J Clin Microbiol. 2003. PMID: 12734210 Free PMC article.
-
Detection of PCR-amplified fungal DNA by using a PCR-ELISA system.Med Mycol. 1998 Oct;36(5):275-9. doi: 10.1080/02681219880000441. Med Mycol. 1998. PMID: 10075496
-
Molecular diagnosis and epidemiology of fungal infections.Med Mycol. 1998;36 Suppl 1:249-57. Med Mycol. 1998. PMID: 9988514 Review.
-
Towards a molecular diagnosis of invasive aspergillosis and disseminated candidosis.FEMS Immunol Med Microbiol. 2005 Sep 1;45(3):361-8. doi: 10.1016/j.femsim.2005.05.012. FEMS Immunol Med Microbiol. 2005. PMID: 16054349 Review.
Cited by
-
A novel extraction method combining plasma with a whole-blood fraction shows excellent sensitivity and reproducibility for patients at high risk for invasive aspergillosis.J Clin Microbiol. 2012 Aug;50(8):2585-91. doi: 10.1128/JCM.00523-12. Epub 2012 May 16. J Clin Microbiol. 2012. PMID: 22593600 Free PMC article.
-
Rapid differentiation of Aspergillus species from other medically important opportunistic molds and yeasts by PCR-enzyme immunoassay.J Clin Microbiol. 2004 Aug;42(8):3495-504. doi: 10.1128/JCM.42.8.3495-3504.2004. J Clin Microbiol. 2004. PMID: 15297489 Free PMC article.
-
Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis.J Clin Microbiol. 2002 Jun;40(6):2224-7. doi: 10.1128/JCM.40.6.2224-2227.2002. J Clin Microbiol. 2002. PMID: 12037092 Free PMC article.
-
Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk.Appl Environ Microbiol. 2004 Aug;70(8):4561-8. doi: 10.1128/AEM.70.8.4561-4568.2004. Appl Environ Microbiol. 2004. PMID: 15294786 Free PMC article.
-
Multicenter comparison of serum and whole-blood specimens for detection of Aspergillus DNA in high-risk hematological patients.J Clin Microbiol. 2013 May;51(5):1445-50. doi: 10.1128/JCM.03322-12. Epub 2013 Feb 20. J Clin Microbiol. 2013. PMID: 23426930 Free PMC article.
References
-
- Buchman T G, Rossier M, Merz W G, Charache P. Detection of surgical pathogens by in vitro DNA amplification. Part 1. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery. 1990;108:338–347. - PubMed
-
- Burgener-Kairuz P, Zuber J P, Jaunin P, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. - PMC - PubMed
-
- Cone R W, Hobson A C, Brown Z, Ashley R, Berry S, Ishak L, Winter C, Corey L. Frequent reactivation of genital herpes simplex viruses among pregnant women. JAMA. 1994;272:792–796. - PubMed
-
- Crampin A C, Matthews R C. Application of the polymerase chain reaction to the diagnosis of candidosis by amplification of an HSP90 gene fragment. J Med Microbiol. 1993;39:233–238. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

