The human immunodeficiency virus type 1 Tat protein up-regulates the promoter activity of the beta-chemokine monocyte chemoattractant protein 1 in the human astrocytoma cell line U-87 MG: role of SP-1, AP-1, and NF-kappaB consensus sites
- PMID: 10644332
- PMCID: PMC111637
- DOI: 10.1128/jvi.74.4.1632-1640.2000
The human immunodeficiency virus type 1 Tat protein up-regulates the promoter activity of the beta-chemokine monocyte chemoattractant protein 1 in the human astrocytoma cell line U-87 MG: role of SP-1, AP-1, and NF-kappaB consensus sites
Abstract
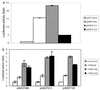
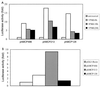
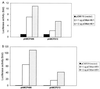
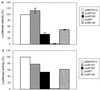
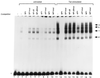
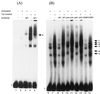
It has been shown that the human immunodeficiency virus type 1 (HIV-1) Tat protein can specifically enhance expression and release of monocyte chemoattractant protein 1 (MCP-1) from human astrocytes. In this study, we show evidence that Tat-induced MCP-1 expression is mediated at the transcriptional level. Transient transfection of an expression construct encoding the full-length Tat into the human glioblastoma-astrocytoma cell line U-87 MG enhances reporter gene activity from cotransfected deletion constructs of the MCP-1 promoter. HIV-1 Tat exerts its effect through a minimal construct containing 213 nucleotides upstream of the translational start site. Site-directed mutagenesis studies indicate that an SP1 site (located between nucleotides -123 and -115) is critical for both constitutive and Tat-enhanced expression of the human MCP-1 promoter, as mutation of this SP1 site significantly diminished reporter gene expression in both instances. Gel retardation experiments further demonstrate that Tat strongly enhances the binding of SP1 protein to its DNA element on the MCP-1 promoter. Moreover, we also observe an increase in the binding activities of transcriptional factors AP1 and NF-kappaB to the MCP-1 promoter following Tat treatment. Mutagenesis studies show that an upstream AP1 site and an adjacent NF-kappaB site (located at -128 to -122 and -150 to -137, respectively) play a role in Tat-mediated transactivation. In contrast, a further upstream AP1 site (-156 to -150) does not appear to be crucial for promoter activity. We postulate that a Tat-mediated increase in SP1 binding activities augments the binding of AP1 and NF-kappaB, leading to synergistic activation of the MCP-1 promoter.
Figures




Similar articles
-
Analysis of the HIV-1 LTR NF-kappaB-proximal Sp site III: evidence for cell type-specific gene regulation and viral replication.Virology. 2000 Sep 1;274(2):262-77. doi: 10.1006/viro.2000.0476. Virology. 2000. PMID: 10964770
-
Cooperative interaction of C/EBP beta and Tat modulates MCP-1 gene transcription in astrocytes.J Neuroimmunol. 2005 Mar;160(1-2):219-27. doi: 10.1016/j.jneuroim.2004.11.009. Epub 2005 Jan 26. J Neuroimmunol. 2005. PMID: 15710476
-
Cytokine induction of monocyte chemoattractant protein-1 gene expression in human endothelial cells depends on the cooperative action of NF-kappa B and AP-1.Eur J Immunol. 1997 May;27(5):1091-7. doi: 10.1002/eji.1830270508. Eur J Immunol. 1997. PMID: 9174597
-
HIV-1 Tat inhibits IL-2 gene transcription through qualitative and quantitative alterations of the cooperative Rel/AP1 complex bound to the CD28RE/AP1 composite element of the IL-2 promoter.J Immunol. 2001 Apr 1;166(7):4560-9. doi: 10.4049/jimmunol.166.7.4560. J Immunol. 2001. PMID: 11254713
-
Stimulus-specific regulation of chemokine expression involves differential activation of the redox-responsive transcription factors AP-1 and NF-kappaB.J Leukoc Biol. 1999 Mar;65(3):291-8. doi: 10.1002/jlb.65.3.291. J Leukoc Biol. 1999. PMID: 10080530 Review.
Cited by
-
Laminins in tumor-derived exosomes upregulated by ETS1 reprogram omental macrophages to promote omental metastasis of ovarian cancer.Cell Death Dis. 2022 Dec 7;13(12):1028. doi: 10.1038/s41419-022-05472-7. Cell Death Dis. 2022. PMID: 36477408 Free PMC article.
-
Human immunodeficiency virus type 1 Tat and methamphetamine affect the release and activation of matrix-degrading proteinases.J Neurovirol. 2004 Feb;10(1):21-8. doi: 10.1080/13550280490261699. J Neurovirol. 2004. PMID: 14982725
-
Gammacoronavirus Avian Infectious Bronchitis Virus and Alphacoronavirus Porcine Epidemic Diarrhea Virus Exploit a Cell-Survival Strategy via Upregulation of cFOS to Promote Viral Replication.J Virol. 2021 Feb 15;95(4):e02107-20. doi: 10.1128/JVI.02107-20. Epub 2020 Nov 25. J Virol. 2021. PMID: 33239458 Free PMC article.
-
Induction of the chemokines interleukin-8 and IP-10 by human immunodeficiency virus type 1 tat in astrocytes.J Virol. 2000 Oct;74(19):9214-21. doi: 10.1128/jvi.74.19.9214-9221.2000. J Virol. 2000. PMID: 10982368 Free PMC article.
-
Coxsackievirus group B type 3 infection upregulates expression of monocyte chemoattractant protein 1 in cardiac myocytes, which leads to enhanced migration of mononuclear cells in viral myocarditis.J Virol. 2004 Nov;78(22):12548-56. doi: 10.1128/JVI.78.22.12548-12556.2004. J Virol. 2004. PMID: 15507642 Free PMC article.
References
-
- Arya S K, Guo C, Josephs S F, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

