Effects of amino acid substitutions at conserved and acidic residues within region 1.1 of Escherichia coli sigma(70)
- PMID: 10613885
- PMCID: PMC94262
- DOI: 10.1128/JB.182.1.221-224.2000
Effects of amino acid substitutions at conserved and acidic residues within region 1.1 of Escherichia coli sigma(70)
Abstract
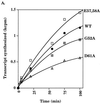
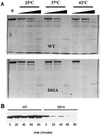
Amino acid substitutions in Escherichia coli sigma(70) were generated and characterized in an analysis of the role of region 1.1 in transcription initiation. Several acidic and conserved residues are tolerant of substitution. However, replacement of aspartic acid 61 with alanine results in inactivity caused by structural and functional thermolability.
Figures


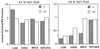
Similar articles
-
Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex.J Mol Biol. 2000 Jun 23;299(5):1217-30. doi: 10.1006/jmbi.2000.3808. J Mol Biol. 2000. PMID: 10873447
-
Analysis of regions within the bacteriophage T4 AsiA protein involved in its binding to the sigma70 subunit of E. coli RNA polymerase and its role as a transcriptional inhibitor and co-activator.J Mol Biol. 2003 Jan 31;325(5):827-41. doi: 10.1016/s0022-2836(02)01307-4. J Mol Biol. 2003. PMID: 12527294
-
Mutations in the 1.1 subdomain of Escherichia coli sigma factor sigma70 and disruption of its overall structure.Eur J Biochem. 1997 Mar 1;244(2):613-8. doi: 10.1111/j.1432-1033.1997.00613.x. Eur J Biochem. 1997. PMID: 9119031
-
Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli.Res Microbiol. 2009 Nov;160(9):667-76. doi: 10.1016/j.resmic.2009.08.014. Epub 2009 Sep 16. Res Microbiol. 2009. PMID: 19765651 Review.
-
Interaction of Escherichia coli sigma 70 with core RNA polymerase.Cold Spring Harb Symp Quant Biol. 1998;63:277-87. doi: 10.1101/sqb.1998.63.277. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384292 Review. No abstract available.
Cited by
-
In vitro properties of RpoS (sigma(S)) mutants of Escherichia coli with postulated N-terminal subregion 1.1 or C-terminal region 4 deleted.J Bacteriol. 2003 Apr;185(8):2673-9. doi: 10.1128/JB.185.8.2673-2679.2003. J Bacteriol. 2003. PMID: 12670993 Free PMC article.
-
Distinct functions of regions 1.1 and 1.2 of RNA polymerase σ subunits from Escherichia coli and Thermus aquaticus in transcription initiation.J Biol Chem. 2012 Jul 6;287(28):23779-89. doi: 10.1074/jbc.M112.363242. Epub 2012 May 17. J Biol Chem. 2012. PMID: 22605342 Free PMC article.
-
The role of region II in the RNA polymerase sigma factor sigma(N) (sigma(54)).Nucleic Acids Res. 2000 Jul 1;28(13):2563-70. doi: 10.1093/nar/28.13.2563. Nucleic Acids Res. 2000. PMID: 10871407 Free PMC article.
-
Formation of intermediate transcription initiation complexes at pfliD and pflgM by sigma(28) RNA polymerase.J Bacteriol. 2001 Nov;183(21):6244-52. doi: 10.1128/JB.183.21.6244-6252.2001. J Bacteriol. 2001. PMID: 11591667 Free PMC article.
-
Rewiring of growth-dependent transcription regulation by a point mutation in region 1.1 of the housekeeping σ factor.Nucleic Acids Res. 2020 Nov 4;48(19):10802-10819. doi: 10.1093/nar/gkaa798. Nucleic Acids Res. 2020. PMID: 32997144 Free PMC article.
References
-
- Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. - PubMed
-
- Gardella T, Moyle H, Susskind M M. A mutant Escherichia coli ς70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. - PubMed
-
- Gopal V, Chatterji D. Mutations in the 1.1 subdomain of Escherichia coli sigma factor ς70 and disruption of its overall structure. Eur J Biochem. 1997;244:613–618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

